New tool reveals DNA structures that influence disease
Posted: 19 December 2019 | Victoria Rees (Drug Target Review) | No comments yet
Researchers have created an algorithm that locates and analyses DNA structures which are linked to the development of certain diseases.

A team of researchers has developed a tool which allows them to study certain DNA structures called topologically associating domains (TADs) which are linked to the progression of disease.
According to the group, from Penn State, US, the newly created algorithm quickly locates and helps to elucidate the complex functions of TADs.
“On your DNA you have genes and regulatory elements, such as promotors and enhancers, that control gene expression, but these two things can be far away from each other,” said Qunhua Li, associate professor of statistics, Penn State. “TADs bring genes together with their regulatory elements, which enables them to begin the process of gene expression.”
The researchers say that disruption of the boundaries that form TADs can expose genes to the wrong regulatory elements and lead to aberrant gene expression that can result in the initiation of diseases such as cancer.
Called the optimised nested TAD (OnTAD), the team’s computational algorithm rapidly identifies the locations of TADs in the genome and enables examination of their internal architectures, important for understanding their biological functions.
By working within a hierarchical view of DNA interactions, the scientists learned that the more densely coiled the DNA is inside TADs, the greater the gene expression. They concluded this is likely due to the fact that more genes are brought into contact with their regulatory elements.
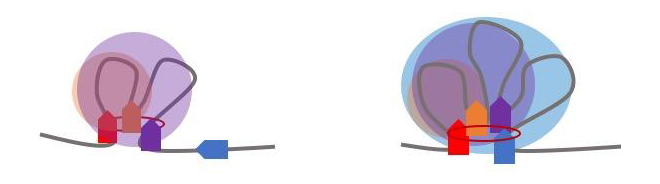
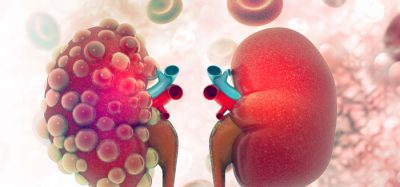
Researchers found that the more densely coiled the DNA is inside TADs (as in the right image compared to the left) the greater the gene expression, likely due to the fact that more genes are brought into contact with their regulatory elements. TADs are areas on the genome within which DNA sequences physically interact with each other more frequently than with sequences outside the TAD (credit: Qunhua Li, Penn State).
“As we better understand how DNA interactions function in normal gene regulation, we can be better prepared to uncover how mutations in DNA can alter those interactions that can lead to incorrect gene expression and influence the development of cancers and other diseases,” said Li.
The researchers note that pre-existing methods have focused solely on identifying the locations of TADs, with little investigation of the biological functions of hierarchical organisation inside the DNA structures.
OnTAD also showed that hierarchical TADs are characterised by more active epigenetic states, which control cell memory and identity.
“These results demonstrate that OnTAD is a powerful tool for revealing different levels of DNA organisation across a genome,” said Li. “It should facilitate improved investigations into the roles of this organisation in gene regulation.”
The results were published in Genome Biology.
Related topics
Disease Research, DNA, Informatics, Technology
Related conditions
Cancer
Related organisations
Penn State
Related people
Qunhua Li