High-throughput protocol for producing tetramer assay probes used to investigate COVID-19
Posted: 21 April 2020 | Hannah Balfour (Drug Target Review) | No comments yet
The developers of a novel method to create immunological assay probes for screening T cells has leveraged their new protocol against COVID-19.
Dendritic cell presenting antigens to T cells with cut away of the T cell receptor and antigen on MHC binding.
Researchers have developed a method that enables them to create libraries containing hundreds of molecular probes for tetramer assays in only days, whereas previous protocols required a week to produce a single probe. The team suggest their process opens up new opportunities for immunological research, development of cancer immunotherapies and assessment of immune responses from patients with COVID-19.
Tetramer assays are an immunological test used to detect and analyse the T cells in a blood sample. T cells each recognise and bind to a specific antigen like a viral protein and bind to them, identifying them to the immune system. However, where antibodies bind directly to antigens on the surface of a pathogen, T cells only bind to antigens expressed on the surface of antigen presenting cells, like macrophages and dendritic cells.
As a result, antibody assays are relatively simple and T-cell assays are more complex; requiring the probes for the latter to be formed of antigens incorporated into a molecular complex that mimics how they are presented in vivo, where they are bound to tetramers of major histocompatibility complex (MHC) proteins.
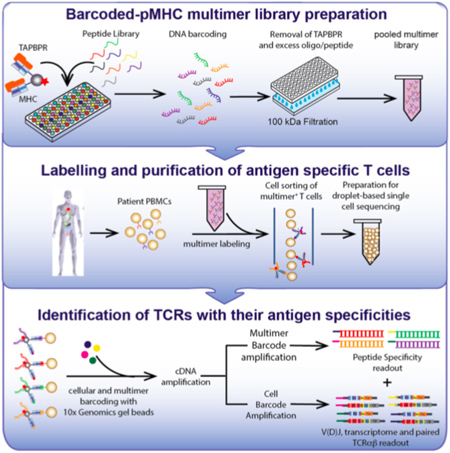
This diagram outlines a workflow for preparing and using a library of peptide-loaded MHC multimers for assessment of T cell repertoires in patient blood samples [credit: Overall et al., Nature Communications, 2020].
Corresponding author of the study published in Nature Communications, Nikolaos Sgourakis, assistant professor of chemistry and biochemistry at the University of California – (UC) Santa Cruz has been studying how protein fragments are selected and bound to MHC proteins in cells for years, a process which requires molecular chaperones. The new protocol is based on his research which indicates that molecular chaperones can eject antigens that have low affinity for the MHC protein tetramers, in favour of antigens with higher affinity.
Sgourakis designed a ‘placeholder’ peptide for use in preparing large quantities of pre-loaded MHC complexes. When incubated with a high-affinity antigen, the placeholder is displaced, a reaction that can be performed in a high-throughput system with large numbers of antigens.
“It’s a force multiplier, enabling us to perform these reactions at high throughput,” Sgourakis said. “A lot of groups are working on similar methodologies, all of which have their pros and cons. This technology has the advantage of using the same system that cells use naturally and we can combine it very elegantly with existing single-cell analytical tools.”
Tetramer assays and COVID-19
The researchers originally used the new method to develop libraries of probes for assessing T-cell responses to neuroblastoma and designing cancer immunotherapies. However, since the emergence of COVID-19, the team have been exploring ways to apply the technology to address the challenges of the novel coronavirus. With a viral infection, there are many different fragments of the viral proteins that an infected cell can display and the researchers say it is important for drug design to determine which of these peptides elicit a strong immune response.
“Based on the coronavirus genome, we can predict all the possible peptides, synthesise them, load them onto MHC tetramers and do a fishing expedition to find which ones are recognised by the T cells in blood samples from patients,” Sgourakis explained. “Certain peptides are immunodominant, ie, they steer the immune response and those are the ones we want to discover so we can potentially use them in a vaccine.”
Sgourakis explained that the tetramer assays could also answer one of the biggest questions surrounding COVID-19: “why is there so much variability in the severity of this disease?”
According to Sgourakis, the approach can be used to compare the T-cell repertoires of different cohorts of patients. It is known that as people age their T-cell repertoire declines, resulting in a weaker immune response to pathogens, which may explain why older people are more vulnerable to COVID-19. Sgourakis said: “We can use this technology to screen patients and see what the gaps are in their T-cell repertoires and maybe use this as a diagnostic for which patients will need more intensive treatment.”
This research was in close collaboration with researchers at the New York Genome Center and the University of Pennsylvania’s Children’s Hospital and Perelman School of Medicine, all US.
Related topics
Analytical Techniques, Assays, Cell-based assays, Disease Research, Drug Development, Immunology, Protein, Proteomics, Research & Development, Screening, T cells
Related conditions
Cancer, Coronavirus, Covid-19, Neuroblastoma
Related organisations
New York Genome Center (NYGC), University of California Santa Cruz, University of Pennsylvania's Children's Hospital, University of Pennsylvania's Perelman School of Medicine
Related people
Nikolaos Sgourakis