Raman technique developed for high-throughput discovery of enzymes
Posted: 10 August 2020 | Victoria Rees (Drug Target Review) | No comments yet
A flow mode Raman-activated cell sorter called FlowRACS has been created by researchers for high-throughput discovery of enzymes and their cell factories.

Chinese scientists have developed a flow mode Raman-activated cell sorter (RACS), called FlowRACS, to support the high-throughput discovery of enzymes and their cell factories, at the precision of one microbial cell.
From the Single-Cell Center in Qingdao Institute of Bioenergy and Bioprocess Technology (QIBEBT), Chinese Academy of Sciences (CAS), the researchers say the mining of useful enzymes and microbial cells that produce them can be very slow and difficult. To address these challenges, the team created FlowRACS.
“This is a flow-mode Raman-activated Cell Sorter that can sort microbial cells and enzymes produced, based on Single-cell Raman Spectrum and in a high-throughput manner,” said Professor Ma Bo, Deputy Director of Single-Cell Center and a senior author of the study.
The researchers note that a single-cell Raman Spectrum can reveal the metabolic function of a cell, such as the kind of oils produced or the rate of carbon dioxide fixed, without destroying the cell. Based on these signals, cells are sorted in RACS. To speed up the sorting, cells can be lined up and moved rapidly through a spectrum detection point one by one. However, such flow-mode RACS can be challenging to use, due to the very small size of microbial cells and the very weak Raman signal.
To improve the process, the scientists applied positive dielectrophoresis (pDEP), which can capture or mobilise particles. This allows trapping fast-moving single microbial cells, processing them precisely yet rapidly through a Raman detection point to recognise those cells that produce particular enzymes or metabolites and then packaging cells into droplets. In the end, the droplets that harbour target cells are sorted. In this pDEP Raman-activated droplet sorting, or pDEP-RADS, the cells do not require culturing, labelling or invasive action in order to be sorted and identified at a high speed.
Diacylglycerol acyltransferases (DGATs) are enzymes that produce triacylglycerols (TAGs). However, the researchers say that traditionally, screening of DGATs has been tedious. For each sample, culture of DGAT-producing cells can take days and subsequent analysis of the cells for oil ingredients can also take days.
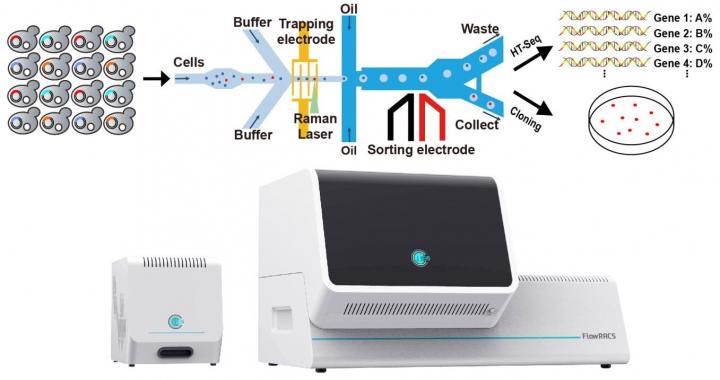
Schematic illustration of the FlowRACS platform and instrument [credit: Wang Xixian].
Using FlowRACS, the researchers screened yeasts that express DGAT candidates for its TAG content and profile, at about 120 cells per minute, equivalent to 120 of the traditional “samples” per minute. They also sorted cells for the degree of unsaturation, a key characteristic of TAG that determines its nutritional value, with about 82 percent accuracy and at 40 cells per minute.
A single run of FlowRACS, which takes just 10 minutes, revealed all previously reported DGAT variants. In contrast, discovering and characterising these enzymes would have either taken months for culture-based methods or simply not been possible due to insufficient sensitivity of fluorescence-activated cell sorting.
“Such culture-free, label-free and non-invasive sorting of enzyme activity in vivo can save time and labour by one to two orders of magnitude, as compared to conventional approaches,” said Dr Wang Xixian, first author of the paper.
“To our knowledge, this is the first demonstration of RACS for enzyme discovery,” added Professor Xu Jian, Director of Single-Cell Center and the other senior author of the study. “The birth of FlowRACS greatly expands the application of RACS. The community is now equipped with a new instrument for the discovery and mining of the ‘biological dark matter’ for not just cells but enzymes.”
The study was published in Science Advances.
Related topics
Disease Research, Enzymes, Protein, Proteomics, Robotics, Screening, Spectroscopy, Technology
Related organisations
Chinese Academy of Sciences (CAS)
Related people
Dr Wang Xixian, Professor Ma Bo, Professor Xu Jian