3D models used to investigate bacteria motility
Posted: 16 May 2023 | Izzy Wood (Drug Target Review) | No comments yet
US researchers created a 3D model of the bacteria: Helicobacter pylori, to better understand its motility, in order to develop new treatments for bacterial infections.


Florida State University’s College of Engineering researchers, US, created a 3D model of Helicobacter pylori (H. pylori) bacteria to better understand its movement, hoping to crack the code governing the organism’s motility and develop alternative treatments for infections, such as strengthening the gastric mucus barrier that stands against the bacteria.
More than 13 percent of Americans have an H. pylori infection, although rates vary with age, race and socioeconomic status. The microorganism uses its corkscrew-like tail to power forward through viscous fluids such as stomach mucus. When it arrives at the epithelium of the stomach wall, it can cause everything from ulcers to cancer.
“People around the world have treated ulcers with antibiotics because antibiotics kill bacteria, but it’s a double-edged sword,” said study co-author Hadi Mohammadigoushki, Associate Professor in the Department of Chemical and Biomedical Engineering. “If we understand how these bacteria move, we can work toward providing other solutions for treatment.”
In the experiments, published by Physical Review Letters, the team placed a model of the bacteria in a high-viscosity polymer gel, an example of what’s called a yield-stress fluid. Those fluids behave as solids under small stresses but flow like liquids beyond a critical stress point.
Next, they used a magnetic field to rotate the 3D model, mimicking the behaviour of the microorganism. Using particle tracking and imaging techniques, the researchers measured the speed of the bacteria and visualised the distribution and density of the fluid flowing around it. The researchers identified two critical thresholds that the bacteria must overcome: the torque needed to rotate the swimming model, and the force needed to propel the model forward.
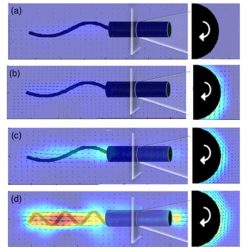

An illustration of the 3D printed model of spiral-shaped bacteria made by researchers at the FAMU-FSU College of Engineering. Using particle tracking and imaging techniques, the researchers measured the speed of the bacteria and visualised the distribution and density of the fluid around it. The diagram shows how the fluid around the model changes as the model spins and moves forward.
(Credit: Hadi Mohammadigoushki)
“We found that if the tail propulsion was too weak, the bacteria remain stuck in the gel,” Mohammadigoushki added. The swimming motions and force that allow H. pylori to move also apply to larger objects, such as earthworms that burrow in the soil, various parasites and more.
“If we understand how the bacteria successfully move to attack our body, we can use that information for whatever we can imagine,” said Kourosh Shoele, Assistant Professor in the Department of Mechanical Engineering.
“In the future, we can design a micro-robot that can deliver a drug to a particular location in the body, in terms of fighting leukemia and other diseases,” Shoele concluded. “Or perhaps we can design tiny robots that use swimming motion and force, like H. pylori, that can dig deep in the sand to explore for water or oil. The possibilities are endless.”
Related topics
3D printing, Targets
Related organisations
Florida State University’s College of Engineering
Related people
Hadi Mohammadigoushki, Kourosh Shoele