Developments in trapped ion mobility spectrometry
Posted: 2 October 2023 | Torsten Müller (Bruker Daltonics) | No comments yet
Torsten Müller from Bruker Daltonics shares how trapped ion mobility spectrometry (TIMS) fundamentally improves immunopeptidomics research and addresses key challenges in this rapidly expanding field.


Immunopeptidomics is a rapidly expanding field at the intersection of immunology and proteomics that focuses on the identification and characterization of peptides presented by major histocompatibility complex (MHC) molecules, also known as the human leukocyte antigen (HLA), on cell surfaces. These peptides are a vital part of the immune system’s display-recognition mechanism that enables T cells to distinguish between self and non-self-antigens, recognizing intruders to initiate immune responses.1 Exploration of the antigenic landscape has gained significant attention for its potential to enhance our understanding of the immune system – and specifically immune targeting at the pre-clinical stage – to help advance our understanding of various diseases. Crucial for personalized cancer immunotherapies and disease pathogenesis insights, MHC-bound peptide identification is facilitated by TIMS-enhanced mass spectrometry (MS).
Immunopeptidomics can identify tumour-specific antigens presented on MHC molecules to provide valuable information for the development of a personalized immunotherapy or vaccine. It allows researchers to identify pathogen-derived peptides presented by infected cells and investigate the immune responses against viral or bacterial infections. Furthermore, immunopeptidomics can play a significant role in understanding autoimmune diseases, biomarker discovery, vaccine and drug development.2
Immunopeptidomics employs advanced MS techniques to identify and quantify MHC-bound peptides to aid vaccine development, autoimmune disease understanding, and cancer immunotherapy development. This involves complex workflows, including cell isolation, MHC peptide enrichment, liquid chromatography MS (LC-MS), and data analysis. Despite many breakthroughs, specific challenges require attention to harness the full potential of immunopeptidomics research to enable the identification of a broad range of MHC-bound peptides.
Challenges in immunopeptidomics
LC-MS-based detection of rare and clinically relevant immunopeptides requires highly sensitive workflows, including sample preparation, data acquisition and data analysis. The low abundance of immunopeptides within complex biological samples presents a challenge in their detection and identification using traditional LC-MS methods. The diversity of major histocompatibility complex (MHC) alleles among individuals and their binding ability to a diverse array of peptides of similar length further complicates the identification of peptides that specifically bind to MHC molecules, necessitating the development of comprehensive MHC-peptide binding databases. Because immunopeptide data is intricate and the source of peptides often unknown, specialized software and computational tools are indispensable in extracting meaningful insights from the data. Overcoming these obstacles holds the potential to unlock valuable insights into immune responses, disease mechanisms and personalized immunotherapies.
Introducing TIMS
TIMS separates ions based on their size and shape, known as the collisional cross section (CCS), influencing their mobility in the gas phase. Adding an extra dimension of separation to LC enhances peak capacity and overcomes traditional challenges. Furthermore, TIMS accumulates and concentrates ions of a specific mass-to-charge (m/z) and mobility, significantly boosting sensitivity and speed. By using a polygon filter in the m/z and mobility space (Figure 1), TIMS allows precise targeting of low-abundance MHC class I and II peptides. Additionally, it facilitates the separation of co-eluting isobaric or nearly isobaric ions using mobility offset, mass aligned (MOMA), improving identification accuracy and confidence.
Parallel accumulation serial fragmentation (PASEF) is a time-of-flight (TOF) MS-based acquisition method enabled by TIMS and employed for fast peptide separation and unambiguous identification. The specificity and high speed of MOMA and PASEF enable confident identification of MHC-bound class I and class II peptides using TIMS, in a single run. Particularly in the field of immunopeptidomics, MHC class I peptides often ionize as single charged species, thereby hampering their identification with traditional workflows. The space charge separation resulting from TIMS allows researchers to readily include single and multiply charged ion species in their analysis. MOMA reduces the interference from co-isolation while the inbuilt polygon filter allows the specific inclusion of precursor ions of interest without compromise.3
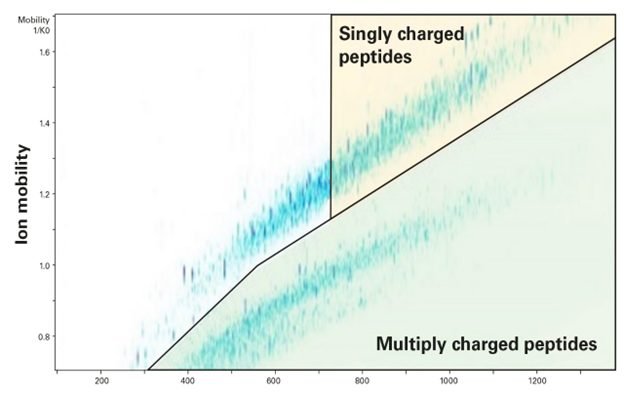

Figure 1. An example of exploiting TIMS and its incorporated polygon filter for precursor ion selection using a timsTOF instrument (Bruker, Billerica, MA) for the analysis of HLA class-I presented peptides.
While for general proteomics, database searching is the preferred approach for peptide identification, this becomes impractical in the field of immunopeptidomics, where enzyme specificity is lacking, and reference databases are unavailable due to the investigation of non-canonical proteins. To address this challenge, a timsTOF optimised de novo sequencing engine (BPS Novor) is integrated into a specialist GPU-driven software to allow for fast, accurate, and precise peptide de novo sequencing of immunopeptidomics data, at a rate of more than 1,000 spectra/second, meaning an hour-long acquisition can be fully de novo sequenced in around two minutes. These exceedingly fast results are obtained without sacrificing accuracy or the availability of database searching of DDA and DIA data on the same platform in real-time.
Conclusion
With new developments in TIMS, PASEF and 4D-proteomics, researchers now can investigate the immunopeptidome with minimal cell amounts, unlocking breakthrough discoveries to contribute to the advancement of personalized medicine. PASEF technology, enabled by TIMS, allows for efficient isolation and detection of ions of interest, offering significant advantages for immunopeptidomics data acquisition and the identification of neoepitopes. The addition of 4D-proteomics offers enhanced peptide separation, discrimination of highly similar peptides, and comprehensive peptidome profiling. Together, these capabilities advance the understanding of complex biological systems and can contribute to immunotherapy advancements.
Author Bio:


Torsten Müller is an LC-MS enthusiast with a strong interest in sample preparation, method optimization, and workflow enhancement within the field of proteomics. He gained valuable experience working in the labs of Hanno Steen in Boston and Ruedi Aebersold in Zurich before completing his PhD at the German Cancer Research Center in Heidelberg, Germany, under the supervision of Prof Jeroen Krijgsveld, where he focused on improving the robustness, reproducibility, and throughput of proteome profiling in clinical specimens.
During his time as a postdoctoral researcher, he successfully implemented an automated workflow based on the SP3 methodology. As Business Development Manager for Proteomics at Bruker, Torsten’s motivation is to make proteomics more accessible to the scientific community at large by fostering collaborations, driving innovation, and empowering researchers to tackle the complexities of the proteome and advance proteomics research.
References:
- Peng X, Woodhouse I, Hancock G, et al. Novel canonical and non-canonical viral antigens extend current targets for immunotherapy of HPV-driven cervical cancer. iScience [Internet]. 2023 [cited 2023 Sep 21];26(3):106101. Available from: https://pubmed.ncbi.nlm.nih.gov/36876126/
- Gravel NH, Nelde A, Bauer J, et al. timsTOF mass spectrometry-based immunopeptidomics refines tumor antigen identification [Internet]. Research Square. 2023 [cited 2023 Sep 21]. Available from: https://www.researchsquare.com/article/rs-2402111/v1
- Gomez-Zepeda D, Arnold-Schild D, Beyrle J, et al. Thunder-DDA-PASEF enables high-coverage immunopeptidomics and identifies HLA class-I presented SarsCov-2 spike protein epitopes [Internet]. Research Square. 2023 [cited 2023 Sep 21]. Available from: https://www.researchsquare.com/article/rs-2625909/v
Related topics
Big Data, Proteomics
Related organisations
Bruker Daltonics
Related people
Torsten Müller (Bruker Daltonics)