Mass spectrometry imaging in drug discovery
Posted: 11 September 2017 | Josephine Bunch (Lead Scientist: NPL), Richard JA Goodwin (Principal Scientist - Drug Safety and Metabolism at AstraZeneca) | No comments yet
Mass spectrometry imaging allows direct measurement of the molecular composition of a sample. It can be used to image the spatial distribution of exogenous drugs and endogenous metabolites simultaneously over the surface of tissue sections or small biopsy, allowing new insights into both compound efficacy and safety during drug discovery.


This label-free imaging modality is influencing pharmaceutical research primarily in pre-clinical discovery and development. However, mass spectrometry imaging is on the cusp of aiding translational research into clinical medicine to enhance patient stratification.
Tackling compound attrition is crucial in reducing the risk of significant financial losses or delay in bringing new medicines to market. It is of paramount importance during drug discovery to understand the efficacy and safety profile of a new therapeutic as soon as possible to allow effective project intervention, redesign of studies, or even cancellation. Efficacy and safety data can allow the findings from failures to positively impact future programmes.
Crucial to making timely project decisions is improved PKPD modelling, which can use translatable efficacy and safety biomarkers to connect the predicted patient population to preclinical animal models. Typically, this is based on quantitative plasma measurements of biomarkers and free-drug levels. These are assumed to be equivalent to tissue concentration. However, there is a significant risk that such assumptions are not always correct. Furthermore, there are many situations where we can assume there will be a discrepancy between plasma and tissue levels. Such situations include poorly vascularised tissues, following localised delivery (inhalation or targeted drug delivery) or the blood-brain barrier. In such cases, using established bioanalytical techniques such as tissue homogenisation, extractions and chromatography coupled to mass spectrometry analysis can help bridge connection between plasma and tissue levels.
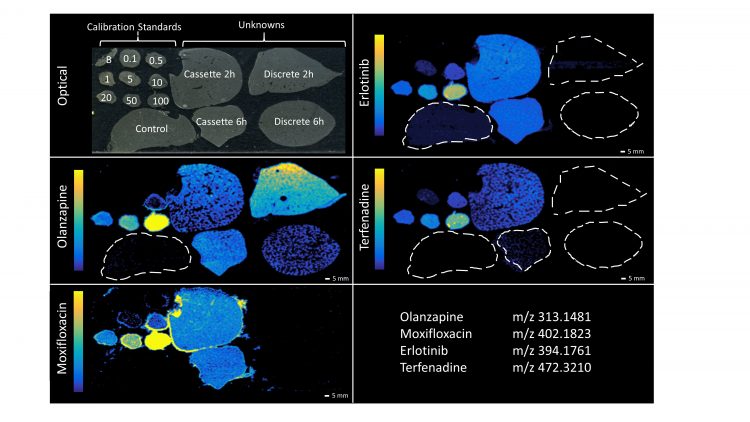

Figure 1: DESI-MSI images of olanzapine, moxifloxacin, erlotinib and terfenadine in rat liver mimetic calibration standards and rat liver ‘unknown’ samples. Poor detection of moxifloxacin, manifested in the compound being detected in all samples and highlighting the halo effect around the top liver mimetic tissue section. Reproduced from Scientific Reports.9
However, such approaches can still have drawbacks, including blood contamination or the dilution of highly localised and low-concentration targets. Furthermore, organs and tissues are not homogeneous but are complex structures with multiple fine structures and cell types in dynamic flux. Therefore, the most insightful analysis of tissues requires the development of technologies that allow the abundance of drugs and biomarkers to be made while also retaining spatial distribution. The situations when plasma measurements alone are imperfect fall into two main categories. The first is when there are non-equilibrium conditions, such as low permeability, limited blood flow or use of tissue targeting delivery systems or sites of local delivery. The second scenario is when free intracellular concentrations cannot be assumed, such as when there is active efflux or uptake or when there could be rapid metabolism in the tissue. Tissue homogenisation bioanalysis has limitations when target biomarkers are unstable (following standard tissue homogenisation) and in understanding biomarker response when addressing complex tissue morphology and multiple cell types.
Mass spectrometry imaging is now delivering impact at all stages of drug discovery. A direct surface sampling of tissue sections, biopsies and even 3D cell cultures, with a variety of desorption/ionisation methods, has now been coupled with a wide range of mass analysers, with ever increasing sensitivity and specificity. It is now even possible to profile and image endogenous metabolites, lipids and proteins from the same sample. This paradigm shift in measurement capability allows us to monitor the interplay between exogenous compounds and biomarkers for compound efficacy and safety. This is a significant step forward from reliance on targeted probes or radiolabelled compounds to monitor tissue exposure and abundance.
Right samples and right technology
MSI can be performed on whole-body tissue sections to investigate possible off-target compound accumulation. However, the majority of MSI is performed on dissected tissues, collected with MSI as the primary analytical endpoint. Careful preparation of modified formalin solutions has been introduced for in situ identification of lipids1 but more typically, tissues should be snap-frozen rather than formalin fixed. Correct sample preparation is crucial to mitigate artificially affecting endogenous biomarkers during tissue collection, processing and samples preparation.2
MSI has been applied to a wide variety of sample types, including cells,3 3D organoids,4 tissues and xenografts, whole rodents, tissue biopsies, tissue microarrays, resection patient samples and even human hair and fingerprints.5,6
A recent tipping point has come as improvements in instrument speed, spatial resolution and sensitivities mean that sufficient data from higher sample numbers, in realistic timescales, means that MSI is now truly impacting project decision making.
Application in drug discovery
The deployment of MSI at any stage of the drug discovery pipeline comes at a significant cost. MS imaging platforms (mass spectrometer and imaging ionization interface) cost between $500,000 and $5m and require expert users and ancillary items (large data storage, sample preparation and processing equipment). Samples are often expensive to generate, eg, pre-clinical studies. Careful project planning is required to ensure that sufficient samples are collected for each measurement, eg, MALDI, SIMS, DESI, pathology and that they are prepared appropriately. MSI is particularly useful for investigating unanticipated histopathology findings and so projects need to balance prospectively, collecting extra samples against increased animal number, husbandry and storage costs. As a result, MSI is still typically used judiciously, to answer specific questions, and is not typically performed as part of every study.
Despite the constraints of publishing sometimes associated with researchers working in the industry, there are abundant examples of how MSI can be used in pharmaceutical R&D. The key areas of pharmaceutical use of MSI are summarised in Table 2.
It is not currently possible to differentiate free drug from protein bound using MSI and this remains a challenge for PKPD modelling. However, quantitative MSI can be used in combination with plasma measurements to partially address this. There are a number of strategies for obtaining quantitative information with MSI. It should be noted that a complete and principled comparison of strategies has not been performed, nor is there full community agreement on the best approach. The simplest method involves preparation of a spiked calibration curve on control tissue, which is analysed alongside the dosed samples. More robust approaches involve the application of a homogeneous coating of a stable-labelled analogue onto the surface of a sample to allow correction for localised ionisation variations caused by tissue matrix effects.7 Quantitation can also be improved through the use of mimetic tissues spiked with target compounds.8 Such mimetics are also useful for quantitation using MSI techniques that samples from larger areas, including liquid micro-junction sampling (LESA – liquid extraction surface analysis).
MSI is most commonly used for analysis of a number of key target organs. For example, imaging drug metabolism in the liver while simultaneously monitoring for early signs of toxicity can be hugely beneficial. Another key target organ that has probably received most MSI analysis is the brain. All the major MSI ionisation methods have been used to map the brain at high spatial resolution, identifying whether compounds are crossing the blood-brain barrier (BBB). By mapping the distribution of blood (by monitoring the haem moiety of haemoglobin) and correlating this to any detected drug, researchers are able to determine if a compound has effectively crossed the BBB or if it is limited to leaky ventricles or just detected in the peripheral blood. This is significantly more powerful than whole brain homogenisation and back calculation for assumed blood contamination. Detection and imaging of neurotransmitters, directly or by using tissue chemical derivatisation, has also been achieved alongside imaging drugs, in the same experiment.10
A recently published example of MSI shows how it can be used to measure and monitor compound absorption in intestinal villi. This is now possible because of the high speed and high spatial resolution MALDI MSI, combined with optimised sample collection and processing to minimise analyte delocalisation or degradation. It shows that MSI can be effectively used for even the most delicate or intricate samples (see Figure 2).11
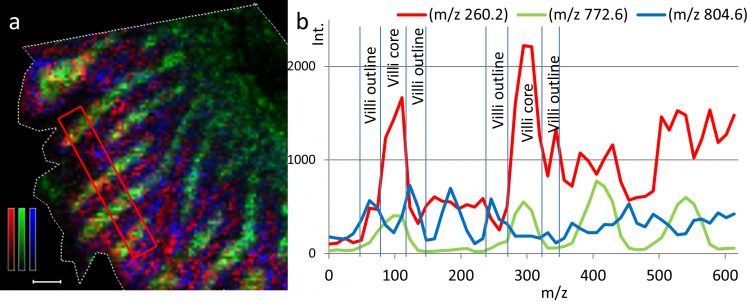

Figure 2: Ion distribution image and transverse profile of propranolol in rat small intestine villi following oral administration. (a) Ion distribution images of propranolol (m/z 260.2) together with two different villi markers; m/z 772.6 represents lamina propria region and m/z 804.6 represents the zone of the epithelial cells of the villi outline. Propranolol is represented by a red monochrome colour scale; scaled to 10% of max intensity, m/z 772.6 is represented in green; scaled to 60% of max intensity, and m/z 804.6 is represented in blue; scaled to 20% of max intensity. Scale bar is 100 µm. (b) The intensities of propranolol (red), m/z 772.6 (green), and m/z 804.6 (blue) are also represented by line graphs. Reproduced from Scientific Reports.11
The ability to perform high spatial resolution MSI is also allowing unprecedented insights into compound distribution and retention following a range of administration routes. Understanding where a compound reaches and remains following inhalation is challenging, even using a radiolabelled compound. Using MSI it is now possible to see the compound’s global distribution. By using high-resolution imaging of compound distribution with the ability to detect endogenous metabolites that define tissue such as blood vessels, bronchi and epithelial layers, it is possible to show retention and absorption at near cellular resolution. Another area of drug delivery that has been analysed by MSI includes mapping the absorption into skin following topical administration.12,13 It has also been used in memetic 3D skin cell cultures.14 Again, due to the recent advances in resolution and sensitivity, MSI has been adopted as a useful tool for analysis of 3D organoids and tissue biopsies.15 This demonstrates how MSI can now be deployed for analysis at near cellular level all the way through to a clinical setting. Using SIMS it is possible to map the distribution of drugs within individual cells.3 The ability to directly measure intracellular distribution of a compound has long been desired by researchers when trying to understand the lack of efficacy or toxicity of a compound. While still not routinely or readily performed, it is clear that the continual advances in MS instrumentation are widening the exciting application areas of MSI.
Looking forward
Over the last decade, MS imaging has evolved from a niche collection of technologies that were slow, insensitive and with unpredictable reproducibility, into a research field that encompasses a huge diversity of application areas and end users. The speed of data acquisition, and the sensitivity and specificity of the mass analysers have reached a stage where they are cost effective and robust enough for non-expert users to start to perform effective imaging studies. The rate of uptake into pharmaceutical research has matched the trajectory of instrument development, with most large pharma having a range of high-performance MSI platforms for project support. By a careful combination of the range of techniques, researchers can collect data to address specific project concerns, while simultaneously collecting data that can be mined more widely.
The metrology and standards required to allow MSI data to be used for regulatory submissions are being developed, led by research at national measurement institutes such as the National Physical Laboratory in the UK. Such work is crucial as MSI expands into clinical trials use where large cohort multi-site studies might be performed. However, there are a number of remaining challenges for the MS imaging community to overcome. A current bottleneck is a lack of widely shared methods for integration and mining of large datasets acquired from multiple imaging platforms, in a range of data formats, at a series of time points.
The challenge can be broken down into two parts. The first, overcoming data integration and vendor specific data formats, is an area where there has already been a drive to open data format. The second, much larger challenge is the amount of information that is now routinely acquired. Each sample might be analysed on three imaging platforms at a range of spatial resolutions, generating hundreds of gigabytes of data from each instrument. This all needs to be stored, processed, mined and compared with other experiments. While imaging data does lend itself to machine learning and artificial intelligence for data mining, demonstrations of these approaches are only just starting to emerge and are not yet widely adopted. None the less, it is clear that MSI presents a phenomenal opportunity for gathering vast amounts of information on the molecular composition of samples in pre-clinical and clinical research and that these data provide powerful new insights to be made into disease biology, progression and prognosis, while also speeding up the development of new medicines.
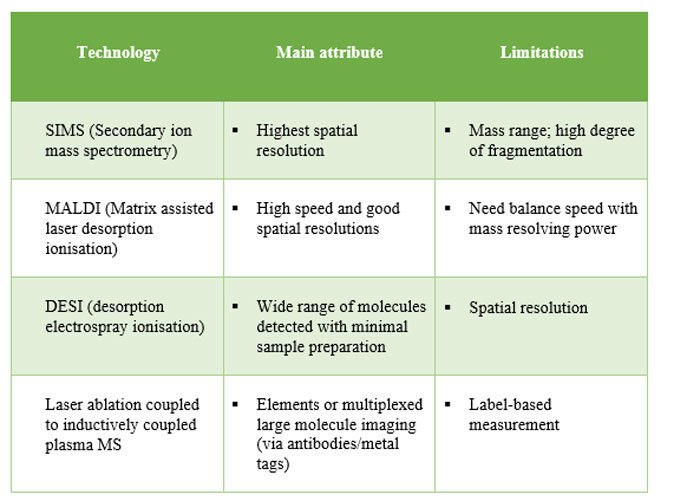

Table 1: Summary of main variations of MS imaging technologies. Use of multiple platforms on same sample set often proves most effective in informing projects.
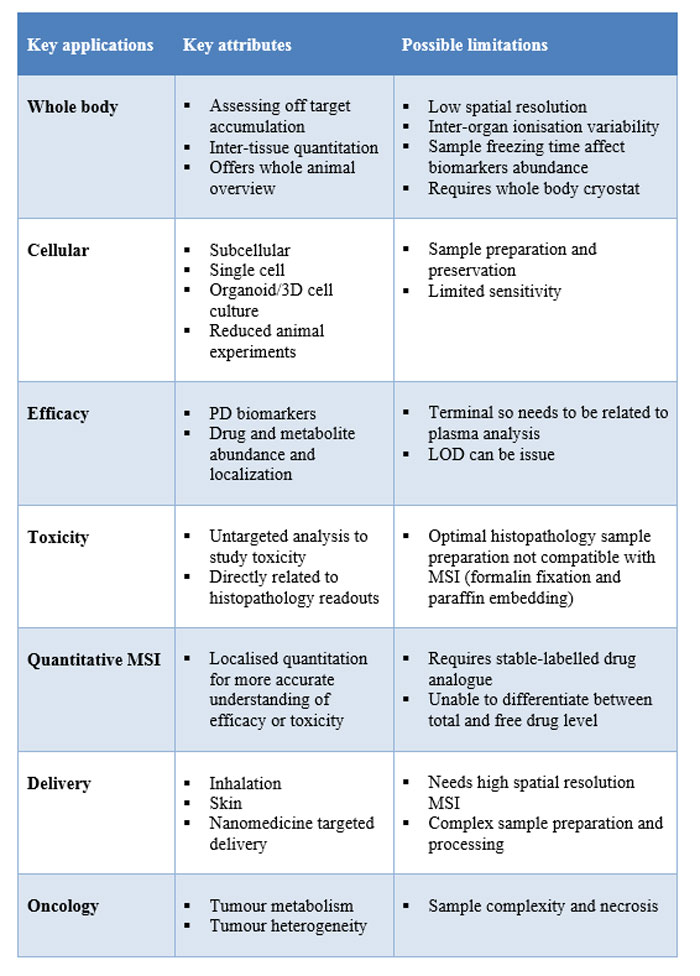

Table 2: Examples of key areas in preclinical drug discovery where MS imaging is often utilised.
About the authors


Dr Richard Goodwin is Principal Scientist at AstraZeneca, Cambridge, UK, where he leads the mass spectrometry imaging group within Drug Safety and Metabolism division. The group supports projects from early discovery through to the clinic. He uses MS imaging to investigate efficacy and safety of new medicines.


Professor Josephine Bunch is a Principal Scientist and Co-Director of the National Centre of Excellence in Mass Spectrometry Imaging (NiCE-MSI) at the National Physical Laboratory, and Chair of Biomolecular Mass Spectrometry at Imperial College London. Prof Bunch is leading the recent £16m Cancer Research UK grand challenge project on mapping tumour metabolism.
References
- Griffiths RL, Sarsby J, Guggenheim EJ, Race AM, Steven RT, Fear J, Bunch J. Formal Lithium Fixation Improves Direct Analysis of Lipids in Tissue by Mass Spectrometry. Analytical Chemistry. 2013;85(15):7146-7153. doi: 10.1021/ac400737z
- Goodwin RJA.Sample preparation for mass spectrometry imaging: Small mistakes can lead to big consequences. Journal of Proteomics. 2012;75(16):4893-4911. doi: http://dx.doi.org/10.1016/j.jprot.2012.04.012
- Passarelli MK, Newman CF, Marshall PS, West A, Gilmore IS, Bunch J, Dollery CT. Single-Cell Analysis: Visualizing Pharmaceutical and Metabolite Uptake in Cells with Label-Free 3D Mass Spectrometry Imaging. Analytical Chemistry. 2015;87(13): 6696-6702. doi: 10.1021/acs.analchem.5b00842
- Liu X, Hummon AB. Mass Spectrometry Imaging of Therapeutics from Animal Models to Three-Dimensional Cell Cultures. Analytical Chemistry. 2015;87(19), 9508-9519. doi: 10.1021/acs.analchem.5b00419
- Cobice DF, Goodwin RJA, Andren PE, Nilsson A, Mackay CL, Andrew R. Future technology insight: mass spectrometry imaging as a tool in drug research and development. British Journal of Pharmacology. 2015;172(13):3266-3283. doi: 10.1111/bph.13135
- Hachmöller O, Aichler M, Schwamborn K, Lutz L, Werner M, Sperling M, Karst U. Element bioimaging of liver needle biopsy specimens from patients with Wilson’s disease by laser ablation-inductively coupled plasma-mass spectrometry. Journal of Trace Elements in Medicine and Biology. 2016;35:97-102. doi: http://dx.doi.org/10.1016/j.jtemb.2016.02.001
- Källback P, Shariatgorji M, Nilsson A, Andrén PE. Novel mass spectrometry imaging software assisting labeled normalization and quantitation of drugs and neuropeptides directly in tissue sections. Journal of Proteomics. 2012;75(16):4941-4951. doi: http://dx.doi.org/10.1016/j.jprot.2012.07.034
- Groseclose MR, Castellino S. A Mimetic Tissue Model for the Quantification of Drug Distributions by MALDI Imaging Mass Spectrometry. Analytical Chemistry. 2013;85(21):10099-10106. doi: 10.1021/ac400892z
- Swales JG, Strittmatter N, Tucker JW, Clench MR, Webborn PJH, & Goodwin RJA Spatial Quantitation of Drugs in tissues using Liquid Extraction Surface Analysis Mass Spectrometry Imaging. Scientific Reports. 2016;6:37648. doi: 10.1038/srep37648.
- Shariatgorji M, Nilsson A, Goodwin RJ, Kallback P, Schintu N, Zhang X, Andren PE. Direct targeted quantitative molecular imaging of neurotransmitters in brain tissue sections. Neuron. 2014;84(4):697-707. doi: 10.1016/j.neuron.2014.10.011
- Nilsson A, Peric A, Strimfors M, Goodwin RJA, Hayes MA, Andrén PE, Hilgendorf C. Mass Spectrometry Imaging proves differential absorption profiles of well-characterised permeability markers along the crypt-villus axis. Scientific Reports. 2017;7:6352. doi: 10.1038/s41598-017-06583-4
- Bunch J, Clench MR, Richards DS. Determination of pharmaceutical compounds in skin by imaging matrix-assisted laser desorption/ionisation mass spectrometry. Rapid Commun Mass Spectrom. 2004;18(24):3051-3060. doi: 10.1002/rcm.1725
- Hart PJ, Clench MR. MALDI-MSI of Lipids in Human Skin. 2017. In LM Cole (Ed.), Imaging Mass Spectrometry: Methods and Protocols (pp.29-36). New York, NY: Springer New York
- Harvey A, Cole LM, Day R, Bartlett M, Warwick J, Bojar R, Clench MR. MALDI-MSI for the analysis of a 3D tissue-engineered psoriatic skin model. Proteomics. 2016;16(11-12):1718-1725. doi: 10.1002/pmic.201600036
- Theiner S, Schreiber-Brynzak E, Jakupec MA, Galanski M, Koellensperger G, Keppler BK. LA-ICP-MS imaging in multicellular tumor spheroids – a novel tool in the preclinical development of metal-based anticancer drugs. Metallomics. 2016;8(4):398-402. doi: 10.1039/C5MT00276A
Related topics
Drug Discovery, Mass Spectrometry
Related organisations
AstraZeneca, National Physical Laboratory
Related people
Josephine Bunch, Richard Goodwin