Phages could help to tackle antimicrobial resistance
Posted: 7 December 2023 | Ellen Capon (Drug Target Review) | No comments yet
Researchers have found what triggers bacteria to start the CBASS immune response to counter infections by phages.
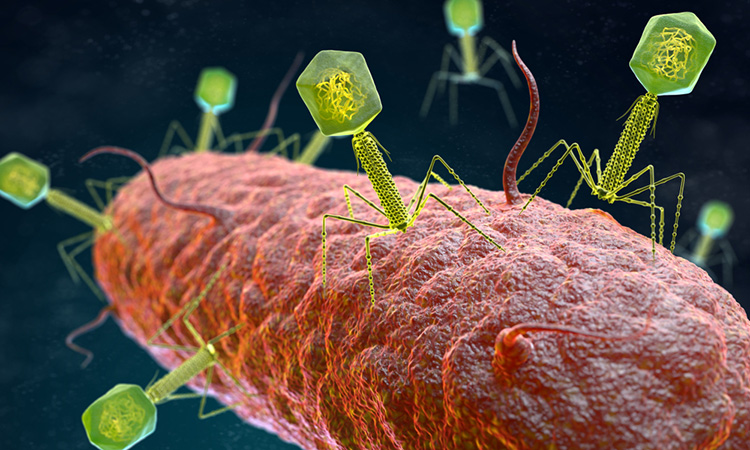

Predatory viruses called phages pose a great threat to bacteria, as they can infiltrate their cells to replicate and take over. Phage have been of interest to scientists as tools to understand fundamental molecular biology, as vectors of horizontal gene transfer and drivers of bacterial evolution, as sources of diagnostic and genetic tools, and as novel therapeutic agents.1
Although bacteria have evolved numerous ways to counter these infections, it was previously unknown how they detect the invading phages. Researchers from the Laboratory of Bacteriology at The Rockefeller University have now found that bacteria sense phages by a defensive response named CBASS (cyclic oligonucleotide-based antiphage signalling system) which detects viral RNA. Dr Luciano Marraffini, head of the lab, said: “Until now, no one has understood what triggers the bacteria to initiate the CBASS immune response.”
Cyclase enzyme
Distantly related domains of life, from eukaryotes, organisms with a membrane-bound nucleus, to prokaryotes, those without such membranes, share core immune functions. These must have evolved in the early existence of life. A viral sensing mechanism that relies on cyclase, a specialised enzyme, is one conserved characteristic.
In animals, this mechanism is named cGAS (cyclic GMP-AMP synthase). In bacteria, cGAS-like cyclases are central parts of the CBASS immune response. Both mechanisms were only discovered in the past decade. Co-author and PhD student Dalton Banh said: “CBASS cyclases are thought to be ancient ancestors of cGAS.”
In an infected animal, cGAS senses viral DNA in cytoplasm whilst in an uninfected organism, DNA is confined in the nucleus. Its presence elsewhere would indicate something is wrong. Bacteria must take a different approach as they do not have nuclei and if CBASS reacted to the presence of DNA, it would cause autoimmunity or the bacterium to attack itself, Banh said. He continued: “CBASS cyclases look a lot like cGAS, so they have to be sensing something. But what, exactly?”
Specific RNA structure
The scientists and their collaborators from Sean Brady’s Laboratory of Genetically Encoded Small Molecules focused on the CBASS system in Staphylococcus schleiferi, a bacterium often located in the mouth of dogs and cats. There have been rare findings of it in humans too.
Dr Marraffini’s has pioneered the study of bacterial defence systems, primarily using CRISPR-Cas. The CRISPR/Cas system has been developed into a new gene-editing tool for the prevention and control of bacterial drug resistance. CRISPR-Cas plays a potentially important role in controlling horizontal gene transfer and limiting the spread of antibiotic resistance.2
His team have many Staph phages available as the lab have used a variety of Staphylococcus strains throughout the years. Banh screened all the phages identified by the defence system. He explained: “This led us to hypothesise that these sensitive phages produced something during infection that triggered activation of CBASS.”
The next stage of the investigation was to test numerous molecules such as DNA, RNA, and proteins, made by the bacterium or the virus. Co-first author Cameron Roberts, a PhD student, conducted this and spoke of the findings: “It was very clearly viral RNA that was generated during infection.” She added: “So instead of sensing a DNA mislocalisation, like cGAS does, CBASS senses a specific RNA structure. This specificity is amazing.”
The novel, hairpin-shaped molecule was named cabRNA for CBASS-activating bacteriophage RNA. It binds to the cyclase surface and initiates the production of a messenger molecule called cGAMP that activates the CBASS immune response.
This parallels how the analogous system operates in humans. Following the detection of viral DNA, cGAS triggers the production of cGAMP, inducing the immune system to produce Type 1 interferons. That antiviral signalling pathway is known as cGAS-STING.
Tackling antimicrobial resistance
Roberts will continue to analyse cabRNA for its characteristics. She said: “Two big questions are how and why the phage generates cabRNA – what is its role?” She continued: “The details of how cabRNA interacts with the CBASS enzyme is also unclear. So solving a structure of the enzyme as it’s bound to the cabRNA would be a huge feat.”
Phages that do not trigger a CBASS response could be useful in the future for tackling antimicrobial resistant bacteria.
Dr Marraffini noted: “Right now, we don’t have the knowledge to predict which phages have the cabRNA and which phages don’t.” He continued: “but if we could do that, we could potentially use those phages to attack bacteria, because they’ve figured out how to slip by this sensing mechanism.”
This study was published in Nature.
References
1 Clokie MRJ, Heaphy S, Letarov AV, Millard AD. Phages in nature. Bacteriophage. 2011 January 1 [2023 November 21]; 1(1):31-45. Available from: https://www.tandfonline.com/doi/full/10.4161/bact.1.1.14942
2 Chen H, Li N, Liang W, Tao S. The Application of the CRISPR-Cas System in Antibiotic Resistance. Infection and Drug Resistance. 2022 September 17 [2023 November 21]; 15(2022):4155-68. Available from: https://www.tandfonline.com/doi/full/10.2147/IDR.S370869
Related topics
Bacteriophages, DNA
Related conditions
Antimicrobial resistance (AMR)
Related organisations
The Rockefeller University