Unique self-activating proteins for drug discovery
Posted: 5 January 2024 | Bishal Parajuli (BellBrook Labs), Dr David P Siderovski (BellBrook Labs), Dr Dustin E Bosch (BellBrook Labs), Joseph Mendez (BellBrook Labs) | No comments yet
Naegleria fowleri infections are rare but almost invariably fatal, with pathologic sequelae spawning its common label as ‘the brain-eating amoeba’. Researchers identified two unique proteins within N. fowleri – self-activating guanine nucleotide-hydrolysing proteins (G proteins) – that present novel targets for drug discovery. This article summarises their work to date, including a high-throughput fluorescence assay of guanine nucleotide hydrolysis for rapid assessment of in silico-predicted small molecule inhibitors.


Naegleria fowleri, a thermophilic free-living amoeba endemic to warm fresh water, is called ‘the brain-eating amoeba’ because it is the causative agent of primary amoebic meningoencephalitis (PAM).1 Intranasal exposure to N. fowleri during recreational watersports is a recurring route to PAM infections (eg, recent ‘splash-pad’ exposures causing paediatric PAM cases)2. Since the mid-1960s, approximately 160 PAM cases have been reported within the US alone, with 98 percent mortality. Researchers and clinicians suggest two key barriers to surviving a N. fowleri infection: delayed positive diagnosis of PAM2 and the apparent lack of effective therapeutic agents targeted specifically to N. fowleri. Recent reviews of N. fowleri cases3 suggest that combinations of antibiotic and antifungal drugs, including azithromycin, amphotericin B, fluconazole, rifampin and miltefosine (the latter originally formulated for leishmaniasis and cancer chemotherapy4), have a minimal impact on survival. The need to identify new drug targets and candidate active pharmaceutical ingredients (APIs) for treating PAM is increasing, as the geographic range of N. fowleri exposure in the US is spreading to more northern climes due to climate change.5
Target identification
In a recent study investigating N. fowleri-specific therapeutic targets,6 the authors used comparative genomics to identify and then biochemically characterise multiple G protein signalling components expressed by this amoeba. Among the signal transduction components identified were 13 N. fowleri G proteins.6 Conventionally, heterotrimeric G protein alpha (Ga) subunits connect agonist-activated G protein-coupled receptors (GPCRs) to intracellular changes in second messenger production (Figure 1a). These conventional Ga proteins bind and hydrolyse guanosine triphosphate (GTP) as part of their activation cycle, with release of the product (GDP) being the rate-limiting step controlled by agonist-bound GPCR-mediated ‘guanine nucleotide exchange factor’ (GEF) activity.7 In contrast to this conventional Ga protein biochemical behaviour, the authors found that two of the thirteen identified N. fowleri Ga proteins (Figure 1b; namely Nf Ga5 and Nf Ga7) exhibit ‘self-activating’ behaviour in vitro:6 extraordinarily rapid spontaneous guanine nucleotide exchange (eg, Figure 1c) coupled with slow GTP hydrolysis. Therefore, these two proteins are predicted to reside mainly in their activated (GTP-bound) state in vivo, irrespective of GPCR influence.
In silico docking
An X-ray crystal structure model of the self-activating G protein Nf Ga7 (Protein Data Bank id 6NE6, 1.70 Å resolution)6 is now being targeted for small molecule inhibitor discovery and development, by exploiting emergent computational tools to identify potential candidate compounds in silico and then test these predicted inhibitors in in vitro biochemical assays. In particular, an ethylene glycol molecule (EDO403) was observed to be stably (and non-covalently) bound within the conventional nucleotide-binding pocket of Nf Ga7 (Figure 2a). This EDO403-binding site has been chosen as the centre of a computational ‘receptor grid’ for the in silico docking of multi-million-element virtual small molecule libraries currently offered by multiple vendors (eg, Enamine, Millipore-Sigma Aldrich Market Select, MolPort, etc). An initial docking screen using Schrödinger’s GLIDE docking algorithm8 as applied to Enamine’s Screening Collection of 4-million+ easily synthesised virtual compounds (average molecular weight ~350 Da; range of 200 ~ 525 Da) provided 144 predicted binders to the EDO403-binding site within Nf Ga7 shown in Figure 2a.
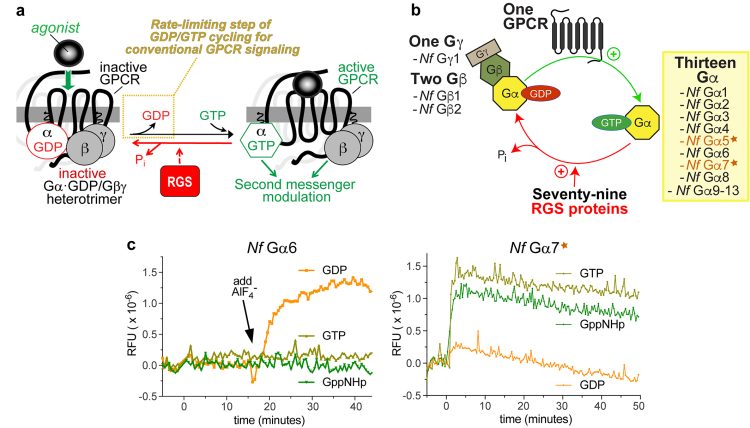

Figure 1
Validating in silico predictions with in vitro testing
To triage the 144 predicted binders, each was purchased, solubilised and tested in vitro for its ability to stabilise (or alternatively destabilise9) the structural integrity of Nf Ga7 upon staged shifts in temperature (ie, within a SYPRO Orange-mediated thermal shift or ‘differential scanning fluorimetry (DSF)’ assay10). The top compounds, as assessed by the absolute value of their resultant Tm shifts (Figure 2b), were then tested in a non-radioactive, steady-state GTPase assay, using BellBrook Labs’ Transcreener® GDP detection reagents, to measure GDP production by purified recombinant Nf Ga7 protein in vitro. The authors had previously pioneered Transcreener® GDP detection for measuring RGS protein enzymatic activity evoking a rate-altered, mammalian Ga point-mutant protein.11 To date, at least one in silico-identified compound (coded Z#1334) has been found to inhibit Nf Ga7 GTPase activity in vitro using the Transcreener® GDP assay (Figure 2c). Current efforts by the authors are now to expand these findings into structure-activity relationship studies with both the Z#1334 chemotype and other chemotypes (as they are discovered), before advancing into testing of more refined leads on free-living amoebic cultures ex vivo.
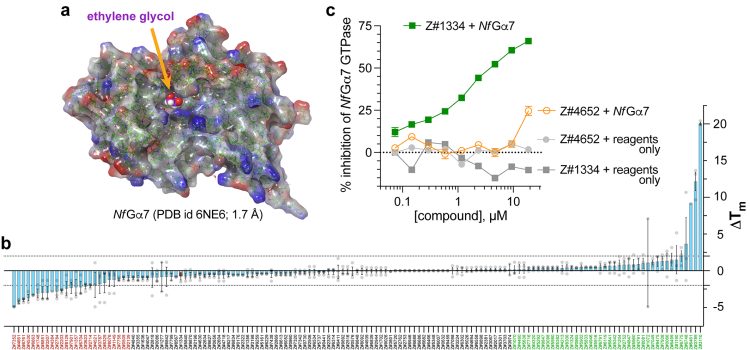

Figure 2
Additional information:
This article summarises ongoing work funded by NIH K08 AI159619 (to D.E.B.), NIH R01 DA048153 (to D.P.S.), and the work reported in: Bosch DE, Jeck WR, and Siderovski DP ‘Self-activating G protein alpha subunits engage seven-transmembrane regulator of G protein signaling (RGS) proteins and a Rho guanine nucleotide exchange factor effector in the amoeba Naegleria fowleri.’ Journal of Biological Chemistry 298(8), 102167 (2022). https://doi.org/10.1016/j.jbc.2022.102167
About the authors


Dr Dustin E Bosch
Dustin completed combined MD and PhD training at the University of North Carolina at Chapel Hill in 2015. His primary focus of PhD study was G protein signalling pathways in the intestinal parasite Entamoeba histolytica with mentorship from Dr David Siderovski. Dustin completed his residency and fellowship training in gastrointestinal and liver pathology at the University of Washington. Since 2021, he has been a physician-scientist in Pathology at the University of Iowa Carver College of Medicine, focused on drug discovery against bacterial and amoebic GI and brain infections.


Joseph obtained his undergraduate training from the University of North Texas at Denton and graduated in 2020 with his bachelor of arts degree in Biology and Chemistry. He is currently a pharmacology & neuroscience PhD candidate, specialising in computational pharmacology, in the Siderovski Lab at the University of North Texas Health Science Center, Fort Worth, TX, USA.


Bishal Parajuli
Bishal received his Bachelor of Arts in Biology and Art from Luther College in 2020. After working for a year as a clinical technologist at Mayo Clinic, he moved to the University of Iowa to immerse himself in a comprehensive educational environment. Since joining the Bosch lab as a research associate at the University of Iowa, he has been actively involved in investigating G-protein signalling in N. fowleri, as well as intestinal bacterial interactions using genetics, biochemistry and structural biology.


Dr David P Siderovski
David received his PhD in Medical Biophysics in 1997 from the University of Toronto, Ontario, Canada, and then completed a four-year industrial postdoctoral position within AMGEN Inc. (Thousand Oaks, CA) in 1999 before returning to academia. He is now a professor of medical education and health systems science at the Texas College of Osteopathic Medicine (TCOM) and a professor of pharmacology & neuroscience at the University of North Texas Health Science Center, Fort Worth, TX, USA.
Figure legends
Figure 1:
Comparison of conventional heterotrimeric G protein alpha subunit biochemical activity with that of the unique Nf Ga7 protein from the brain-eating amoeba N. fowleri. (a) The conventional cycle of GTP binding and GTP hydrolysis by Ga subunits within the standard model of G protein-coupled receptor (GPCR) signal transduction cascade. Agonist-activated GPCRs facilitate the rate-limiting step of Ga subunit guanine nucleotide cycling by exerting guanine nucleotide exchange factor (GEF) activity (highlighted in the yellow dotted box). RGS proteins (Regulators of G protein Signalling) serve to accelerate the intrinsic GTPase activity of Ga subunits (illustrated with the red arrow), but the intrinsic GTPase activity of Ga subunits does not represent the rate-limiting step of GDP/GTP cycling by isolated recombinant Ga subunits in vitro.11 (b) Summary of results from the authors’ N. fowleri comparative genomics study6 that identified multiple G protein signalling components expressed by this amoeba. Among the 13 N. fowleri Ga proteins identified, and subsequently expressed and tested for biochemical activities in vitro,6 two N. fowleri Ga subunits (Nf Ga5 and Nf Ga7; indicated by the star icon) exhibited spontaneous guanine nucleotide exchange in vitro. (c) Conventional behaviour from recombinant purified Nf Ga6 protein was observed when mixed with indicated guanine nucleotides and then monitored spectrophotometrically for any intrinsic tryptophan fluorescence changes indicative of structural conformation alterations characteristic of Ga subunit activation;7 GppNHp = guanylyl imidodiphosphate, a non-hydrolysable GTP analogue. Note no changes in relative fluorescence units (RFUs) were seen for the first 15 minutes, irrespective of guanine nucleotide present. At the indicated time (black arrow), NaF and AlCl3 were added to form the planar anion AlF4–, known to mimic the third phosphate leaving group and move Ga·GDP subunits into an activated conformation.12 (d) In contrast to this conventional behaviour exhibited by Nf Ga6, purified recombinant Nf Ga7 protein was seen7 to move spontaneously into its activated (GTP-bound) conformation upon mixing with either GTP or the non-hydrolysable GTP analogue GppNHp; similar behaviour in this assay was also documented for purified recombinant Nf Ga5.7
Figure 2:
Structural model of the chosen N. fowleri protein target (Nf Ga7) and initial biochemical testing of compounds predicted to interact with the EDO403-binding site of nucleotide-free Nf Ga7. (a) Electrostatic surface rendering of the 1.7 Å-resolution structural model of nucleotide-free Nf Ga7 described by the authors.7 One ethylene glycol molecule (designated within PDB record 6NE6 as ‘EDO403’) is observed in the underlying electron density data to be non-covalently bound within the conventional nucleotide-binding pocket of Nf Ga7 (highlighted in magenta). (b) Results from a SYPRO Orange-based differential scanning fluorimetry (DSF) assay measuring the thermal melt shift (DTm) of Nf Ga7 when co-incubated with the indicated compound (coded on the x-axis) derived from in silico GLIDE docking of Enamine’s 4-million+ Screening Collection into the EDO403-binding site within Nf Ga7. Compounds at the extremes of stabilising (in green) or destabilising (in red) Nf Ga7 in the DSF assay were then selected for subsequent tests in orthogonal, in vitro assays of Nf Ga7 function. (c) Example results from an active small-molecule inhibitor of Nf Ga7, coded Z#1334, when tested in an orthogonal, steady-state Nf Ga7 GTPase assay detected by using fluorescence anisotropy-based BellBrook Labs’ Transcreener® GDP reagents to quantify the GDP produced at 30°C over 120 minutes from GTP substrate (green squares). No background signal was detected upon incubating Z#1334 at indicated concentrations with Transcreener® GDP reagents only (ie, no Nf Ga7 GTPase input). The activity of Z#1334 in inhibiting Nf Ga7 GTPase assay (IC50 of 2.4 mM) appears specific, as an active compound (Z#4652) from a different, RGS-focused drug discovery campaign in the Siderovski lab13 does not detectably inhibit Nf Ga7 GTPase activity, except modestly (~25%) at the highest dose tested (orange open circles).
References
- Ahmad Zamzuri MI, Abd Majid FN, Mihat M, et al. Systematic Review of Brain-Eating Amoeba: A Decade Update. Int J Environ Res Public Health. 2023;20(4). Epub 20230209. doi: 10.3390/ijerph20043021. PubMed PMID: 36833715; PMCID: PMC9964342.
- Eger L, Pence MA. The Brief Case: A Case of Primary Amebic Meningoencephalitis (PAM) after Exposure at a Splash Pad. J Clin Microbiol. 2023;61(7):e0126922. Epub 20230720. doi: 10.1128/jcm.01269-22. PubMed PMID: 37470480; PMCID: PMC10358179.
- Cope JR, Conrad DA, Cohen N, et al. Use of the Novel Therapeutic Agent Miltefosine for the Treatment of Primary Amebic Meningoencephalitis: Report of 1 Fatal and 1 Surviving Case. Clin Infect Dis. 2016;62(6):774-6. Epub 20151217. doi: 10.1093/cid/civ1021. PubMed PMID: 26679626; PMCID: PMC4775347.
- Eibl H, Unger C. Hexadecylphosphocholine: a new and selective antitumor drug. Cancer Treat Rev. 1990;17(2-3):233-42. doi: 10.1016/0305-7372(90)90053-i. PubMed PMID: 2272038.
- Gharpure R, Gleason M, Salah Z, et al. Geographic Range of Recreational Water-Associated Primary Amebic Meningoencephalitis, United States, 1978-2018. Emerg Infect Dis. 2021;27(1):271-4. doi: 10.3201/eid2701.202119. PubMed PMID: 33350926; PMCID: PMC7774533.
- Bosch DE, Jeck WR, Siderovski DP. Self-activating G protein alpha subunits engage seven-transmembrane regulator of G protein signaling (RGS) proteins and a Rho guanine nucleotide exchange factor effector in the amoeba Naegleria fowleri. The Journal of biological chemistry. 2022;298(8):102167. Epub 20220620. doi: 10.1016/j.jbc.2022.102167. PubMed PMID: 35738399; PMCID: PMC9283941.
- Kimple AJ, Bosch DE, Giguere PM, Siderovski DP. Regulators of G-protein signaling and their Galpha substrates: promises and challenges in their use as drug discovery targets. Pharmacol Rev. 2011;63(3):728-49. Epub 20110707. doi: 10.1124/pr.110.003038. PubMed PMID: 21737532; PMCID: PMC3141876.
- Friesner RA, Murphy RB, Repasky MP, et al. Extra precision glide: docking and scoring incorporating a model of hydrophobic enclosure for protein-ligand complexes. J Med Chem. 2006;49(21):6177-96. doi: 10.1021/jm051256o. PubMed PMID: 17034125.
- Charveriat M, Reboul M, Wang Q, et al. New inhibitors of prion replication that target the amyloid precursor. J Gen Virol. 2009;90(Pt 5):1294-301. Epub 20090304. doi: 10.1099/vir.0.009084-0. PubMed PMID: 19264641.
- Bosch DE, Abbasian R, Parajuli B, et al. Structural disruption of Ntox15 nuclease effector domains by immunity proteins protects against type VI secretion system intoxication in Bacteroidales. mBio. 2023;14(4):e0103923. Epub 20230622. doi: 10.1128/mbio.01039-23. PubMed PMID: 37345922; PMCID: PMC10470768.
- Bosch DE, Zielinski T, Lowery RG, Siderovski DP. Evaluating modulators of “Regulator of G-protein Signaling” (RGS) proteins. Curr Protoc Pharmacol. 2012;Chapter 2:Unit2 8. Epub 2012/03/03. doi: 10.1002/0471141755.ph0208s56. PubMed PMID: 22382998; PMCID: PMC3306263.
- Phillips WJ, Cerione RA. The intrinsic fluorescence of the alpha subunit of transducin. Measurement of receptor-dependent guanine nucleotide exchange. Journal of Biological Chemistry. 1988;263(30):15498-505. doi: https://doi.org/10.1016/S0021-9258(19)37616-1.
- Siderovski DP, Agogo-Mawuli PS. Rapidly transitioning from in silico hits to leads on ‘undruggable’ pain targets. Drug Target Review. 2022;9(4):14-6.
Related topics
Hit-to-Lead
Related organisations
BellBrook Labs





