Ingenious AI method for improved target identification
Posted: 12 January 2022 | Mandy Parrett (Drug Target Review) | No comments yet
A research team based at Skoltech in Russia has developed an artificial intelligence-driven solution for highly accurate detection of efficacious binding sites to expedite drug discovery.
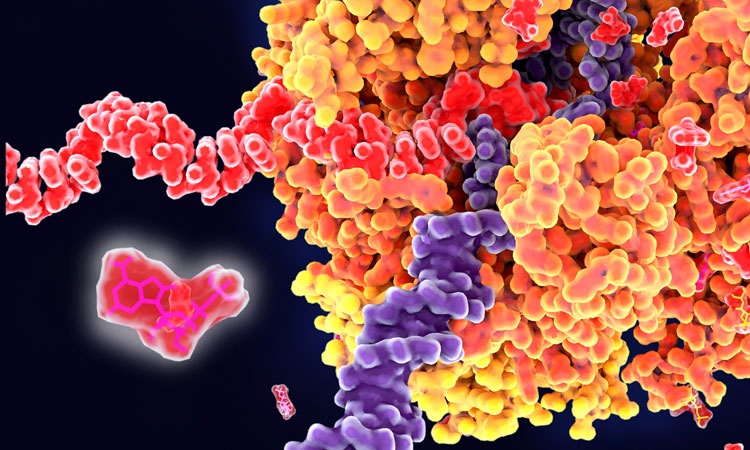

Traditional approaches often target proteins as a means of therapeutic intervention, due to their wide-ranging functionality in our bodies. However, in light of our evolving insight regarding the extensive role played by RNA in cells, scientists now recognise the value of targeting RNA and potentially DNA sequences instead.
Although approximately 85 percent of the genome (DNA) is transcribed into RNAs, only a small percentage of this RNA encodes functional proteins. The rest are noncoding RNAs, which carry out a myriad of functions such as activating or inactivating certain genes or folding into various shapes to fulfil other functions. These new shapes are known as conformations and can have a pathological impact, hence they are seen as potential targets.
As a result of new research, the iMolecule group from Skoltech, as reported in Nucleic Acid Research: Genomics and Bioinformatics, has developed an artificial intelligence-driven solution that uses data on the structure of RNA or DNA molecules to identify sites on them where interaction with potential drug candidates can occur. Information about these binding sites enables pharma companies to discover new medications with greater accuracy and efficiency because it accounts for how the shape assumed by a nucleic acid molecule affects which binding sites are exposed.
“Nucleic acids — DNA and RNA — can participate in signalling, for example, and we could target that or any other process they are involved in. This could be a promising strategy for undruggable protein targets, for example, disordered proteins or proteins that lack convenient binding sites,” Skoltech Assistant Professor Petr Popov, the principal investigator of the study, said. “And then there’s also pathogenic RNA foreign to the body, for example in viruses, such as SARS-CoV-2 or HIV.”
To unlock the potential of these identified drug targets, pharmacologists screen large libraries of chemical compounds to see which of them interact with nucleic acids and pinpoint the precise binding sites.
“We created this new solution by adapting our prior work with proteins,” Popov explained. “Nucleic acid three-dimensional structures are encoded as high-dimensional tensors. Once this is done, a computer vision algorithm ‘looks’ at the tensors and highlights the areas in the structure that it thinks could serve as binding sites. After the conformation and the binding site have been detected, a more focused drug discovery campaign can be initiated. So our work is a small step toward rational drug discovery in contrast to the blind screening, which becomes less reliable with growing chemical libraries.”
However, conformations are complex in their behaviour and can twist and change their shapes, altering which binding sites are exposed. This has led to unreliability in drug effectiveness.
“Most earlier methods only worked with RNA, and specifically, with a single chain. Ours works with DNA and with two or more chains. We can even see additional sites that arise when multiple molecules become entangled,” Igor Kozlovskii, a Skoltech PhD student and the first author of the paper, said.
“A great example of what makes working with methods that ignore conformation problematic is the dominant type of HIV,” he continued. “It has an RNA region targeted by many agents. But even though the nucleic acid sequence is the same, when that molecule changes conformation, this is known to have an effect on which agents work or don’t. Our neural network predictions actually reproduce this effect, which means they are reliable.”
Related topics
Analysis, Artificial Intelligence, Drug Discovery, Drug Discovery Processes, Drug Targets, RNAs, Targets
Related organisations
Nucleic Acid Research: Genomics and Bioinformatics, Skolkovo Institute of Science and Technology, Skoltech
Related people
Igor Kozlovskii (Skoltech), Professor Petr Popov (Skoltech)