Disabling the SETD1B enzyme halts leukaemia cell growth
Posted: 2 July 2025 | Drug Target Review | No comments yet
Japanese researchers have identified the epigenetic enzyme SETD1B as a key driver of aggressive acute myeloid leukaemia (AML) – which could lead to new treatment strategies targeting the cancer’s underlying biology in the future.

Researchers in Japan have uncovered a new therapeutic target that could transform how aggressive forms of acute myeloid leukaemia (AML) are treated, especially in patients with FLT3-ITD mutations – a group historically associated with poor outcomes and frequent relapses. The study, published in Leukemia, identifies the epigenetic enzyme SETD1B as a critical promoter of leukaemia growth via activation of the MYC oncogene – a gene that helps to control how cells grow, divide and survive.
An urgent need for new AML therapies
Acute myeloid leukaemia is a fast-progressing cancer of the blood and bone marrow. While advances in chemotherapy and targeted therapies have improved outcomes for some patients, many AML cases – particularly those with genetic mutations like FLT3-ITD – are still resistant to treatment.
A research team from Chiba University in Japan wanted to understand what drives these aggressive leukaemia’s. Their work looked at the role of SETD1B, an epigenetic regulator, in supporting uncontrolled cancer cell growth.
AI-powered drug discovery: Accelerating the development of life-saving therapies
18 September 2025 | 14:00PM BST | FREE Webinar
Join this webinar to learn how AI is accelerating early-stage drug discovery and improving target identification, practical strategies for applying AI effectively within your organisation and to ask your questions to our industry expert! Dr Remco Jan Geukes Foppen will share practical insights into how AI is being applied across the pharmaceutical sector, helping teams move faster and make better-informed decisions. With experience spanning data management, image analysis, bioinformatics, and machine learning in clinical research, he brings both deep technical expertise and strategic understanding of real-world challenges.
Register Now – It’s Free!
“Patients with AML, especially with the FLT3-ITD mutation, often respond poorly to current therapies. Our findings are that the epigenetic regulator SETD1B protein supports aggressive cell proliferation in AML by promoting oncogenic MYC expression,” said Lead Professor of the study, Takayuki Hoshii.
Epigenetics and AML: the role of SETD1B
The study focused on a biochemical process called H3K4me3 (histone H3 lysine 4 trimethylation). This process modifies how DNA is packaged in cells and regulates gene activity without altering the genetic code. In previous research, high H3K4me3 levels were linked to mixed-lineage leukaemia-rearranged (MLL-r) AML. However, the connection between H3K4me3 and FLT3-ITD AML remained poorly understood.
Using genetic screening techniques, the researchers pinpointed SETD1B as the gene responsible for this epigenetic modification. By adding methyl groups to histone proteins, SETD1B facilitates increased activity of MYC – a gene known to drive cancer when overexpressed.
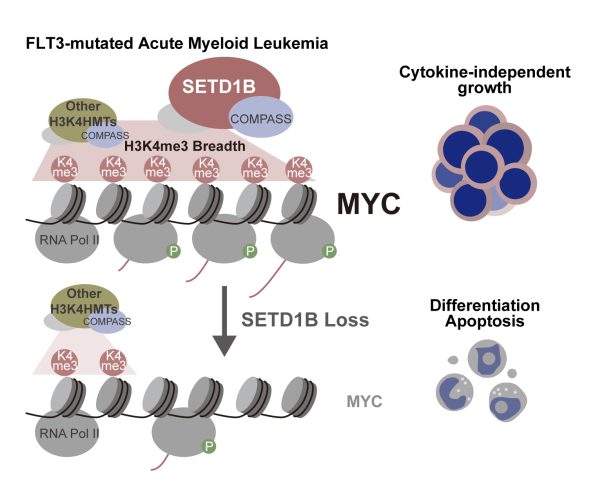

This model illustrates how SETD1B promotes the expansion of H3K4me3 epigenetic marks and upregulates MYC expression, driving cytokine-independent cell growth in FLT3-mutated AML. Credit: Dr. Takayuki Hoshii from Chiba University, Japan
Blocking SETD1B slows cancer growth
To test how SETD1B influences AML progression, the team used CRISPR gene-editing to delete its catalytic domain – the part of the protein responsible for making the H3K4me3 modification. This allowed them to see what happened when SETD1B’s epigenetic function is turned off.
Their results showed AML cells with FLT3-ITD or NrasG12D mutations significantly reduced growth when SETD1B was disrupted.
Their results showed AML cells with FLT3-ITD or NrasG12D mutations significantly reduced growth when SETD1B was disrupted. At the same time, MYC pathway genes were also downregulated, indicating that the cancer cells could no longer rely on MYC to fuel their expansion.
“The breadth of H3K4me3 is crucial for transcriptional consistency, and MYC expression appears highly dependent on both the quality and quantity of transcriptional elongation,” explains Dr Hoshii. “Understanding SETD1B’s role in maintaining this epigenetic mark is critical for developing biomarkers and therapies for leukaemia subtypes and other MYC-driven cancers.”
A foundation for future therapies
To further confirm SETD1B’s influence, the researchers reintroduced the Myc gene into leukaemia cells lacking SETD1B. While this partially restored cell growth, it did not fully reverse the effects – meaning SETD1B controls other critical processes beyond MYC activation.
The study’s findings offer new opportunities for targeted therapies. One potential candidate is Chaetocin, a known inhibitor of related methyltransferase enzymes. While not yet specific to SETD1B, it could be used as a starting point for developing drugs that selectively disrupt SETD1B’s function.
By measuring SETD1B activity, clinicians may also be able to identify which patients are most likely to benefit from such treatments in the future.
A new path forward for AML patients
As scientists continue to explore treatments for aggressive leukaemia’s, targeting the epigenetic machinery of cancer cells offers a new and promising strategy. This latest research not only identifies a new vulnerability in FLT3-ITD AML but also highlights the wider importance of epigenetic regulation in cancer progression.
Related topics
Cancer research, Drug Discovery, Drug Targets, Epigenetics, Molecular Biology, Oncology, Translational Science
Related conditions
acute myeloid leukemia (AML)
Related organisations
Chiba University
Related people
Takayuki Hoshii (Professor at Chiba University)