Computational model used to predict drug targets against SARS-CoV-2
Posted: 26 November 2020 | Victoria Rees (Drug Target Review) | No comments yet
A model of a human lung cell has been used to understand how SARS-CoV-2 uses host cell processes to reproduce, revealing drug targets.

A computational model of a human lung cell has been used to understand how SARS-CoV-2 draws on human host cell metabolisms to reproduce. According to the researchers, this study helps understand how the virus uses the host to survive and enable drug predictions for treating the virus to be made. The study was conducted by researchers at the University of Warwick, UK.
The team highlight that viruses rely on their host to survive. A crucial step of lifecycle is the synthesis of the virus particles within the host cell, so understanding this process is key to finding ways to prevent the virus from surviving.
Using a computer model of a human lung cell metabolism, the scientists captured the stoichiometric amino and nucleic acid requirements of SARS-CoV-2, the virus that causes COVID-19. The model revealed host-based metabolic perturbations inhibiting SARS-CoV-2 reproduction, highlighting reactions in the central metabolism, as well as amino acid and nucleotide biosynthesis pathways. The researchers found that only few of these metabolic perturbations are able to selectively inhibit virus reproduction.
The team also note that some of the catalysing enzymes of such reactions have demonstrated interactions with existing drugs, which can be used for experimental testing of the presented predictions using gene knockouts and RNA-interference techniques.
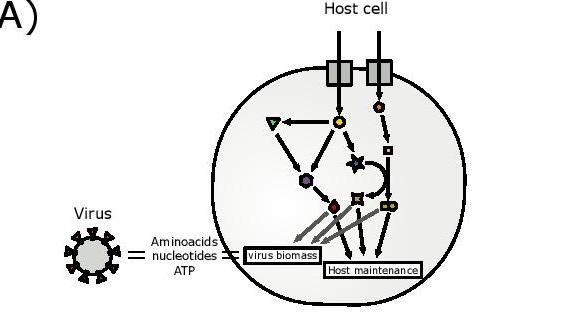

Schematic representation of the integrated host-virus metabolic modelling approach used in the article. The biomass composition of SARS-CoV-2 is estimated based on genomic and structural informations and then embedded in the metabolic network model of the host cell. This model is then used to predict the metabolic fluxes supporting virus production and effects of perturbations [credit: University of Warwick].
Professor Orkun Soyer, one of the researchers, said: “We have created a stoichiometric biomass function for the COVID-19-causing SARS-CoV-2 virus and incorporated this into a human lung cell genome scale metabolic model. We then predicted reaction perturbations that can inhibit SARS-CoV-2 reproduction in general or selectively, without inhibiting the host metabolic maintenance. The predicted reactions primarily fall onto glycolysis and oxidative phosphorylation pathways and their connections to amino acid biosynthesis pathways.”
Another researcher, Dr Hadrien Delattre, added: “Together, these results highlight the possibility of targeting host metabolism for inhibition of SARS-CoV-2 reproduction in human cells in general and in human lung cells specifically. More research needs to be carried out to explore SARS-CoV-2 infected cells and their metabolism, however the model developed here by the researchers can be used as a starting point for testing out specific drug predictions.”
The study was published in the journal Life Science Alliance.
Related topics
Disease Research, Drug Targets, Informatics, Research & Development
Related conditions
Covid-19
Related organisations
Warwick University
Related people
Dr Hadrien Delattre, Professor Orkun Soyer