Antibody library screening: high throughput strategies
Posted: 10 June 2020 | Emma Cummins - Centre for Drug Research and Development Canada, Gregorio Aversa - Abimmune Technologies Inc., Ismael Samudio - Centre for Drug Research and Development Canada | No comments yet
As the number of antibody discovery platforms and formats expands, various characteristics must be rigorously screened for and taken into consideration when deciding which hits to progress to successful development. The earlier in the antibody discovery pathway the assessment is carried out, the greater the hit enrichment and selection will be, leading to more controlled attrition along the pipeline. In this review we will discuss phage display antibody library screening challenges, and commonly used high-throughput screening methods that can be applied to solve them.

Phage display has many advantages, including the ability to drive selection towards desired characteristics such as affinity, specificity and cross-reactivity. However, being a bacterial system displaying scFvs, not full length IgGs, it has limitations needing to be overcome for early stage assays. Following several rounds of selection on antigen, each individual clone is produced as a periplasmic prep (periprep) for primary screens. Peripreps can be produced in high numbers (and repeatedly from master glycerol stocks), enabling screening of thousands of clones per day in antigen-binding assays including ELISA and multiplexed flow cytometry. Although the concentration of periprep is unknown, yes / no identification of binders often provides sufficient data to progress hits to subsequent screens.
However, further information, including ligand-receptor blocking function, may be desirable. Homogenous time-resolved fluorescence (HTRF) is a highly sensitive, high-throughput assay. HTRF can be used to assess the ability of scFv peripreps to block protein-protein interactions via energy transfer measurements, enabling the identification of functional hits that can be compared to other antibodies, for affinity and epitope binding.1,2 To further rank hits, scFvs may be purified from peripreps using simple, rapid, high-throughput immobilised metal affinity chromatography (IMAC) methods,3 yielding enough protein to characterise in numerous assays including binding, neutralisation and even 72h proliferation assays. Figure 1 shows a primary binding screen using single shot peripreps in multiplexed flow cytometry, and the correlation with a number of purified scFvs tested in the same assay.
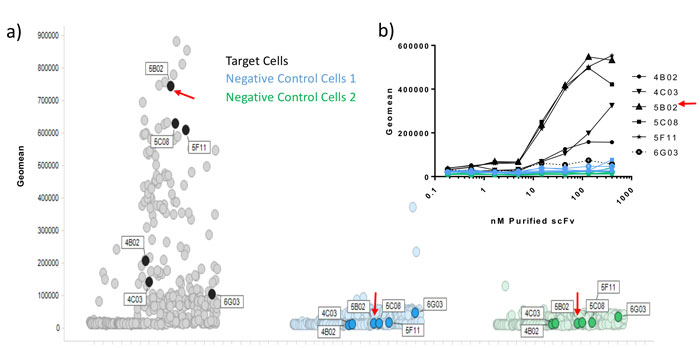

Figure 1: Correlation of periprep and purified scFv data in a multiplexed flow cytometry assay. Following several rounds of selection on target expressing cells, and de-selection on irrelevant negative control cells (1 and 2), individual scFv peripreps were screened by flow cytometry in a multi-plexed screen, where the 2 negative control lines were pre-labelled with barcoding dyes, and then mixed with unlabeled target cells. A large number of hits bound to target cells and not to negative control cells. 5 target cell hits and 1 non-specific binder (6G03), were subsequently purified by high-throughput IMAC. Binding was confirmed by flow cytometry (inset graph) and ranking correlated with the periprep single shot data.
Data analysis comparing ranking of scFv vs IgG typically indicates a general trend (data not shown), enabling confident selection of scFvs, however, early biologically-relevant format screening is advisable. ScFvs tend to form a heterogeneous mixture of monomer / dimer, therefore affinity measurement requires reformatting to IgG. Thus, the ability to work directly with full-length IgGs is an advantage for yeast or mammalian display platforms. Interestingly, an E.coil-mammalian dual expression system using a scFv.Fc vector (pSplice) has been reported.4 This system enables phage library screening in svFv. Fc format, adding functionality for rapid in-depth secondary screening, after direct transfection and expression in mammalian cell lines. Epitope binning is key to ranking large antibody panels, by determining which antibodies may have similar epitope and therefore function. Abdiche et al.5, suggested to ‘bin the fridge’, as the epitope is an innate property that cannot be engineered, therefore a critical feature of hit selection, while antibody affinity and stability can be optimised. High-throughput binning has been made more achievable with biosensors such as the Octet or Carterra LSA platforms, which enable simultaneous binning of 384 antibodies from supernatant.6 A typical phage display screening workflow is shown in Figure 2.
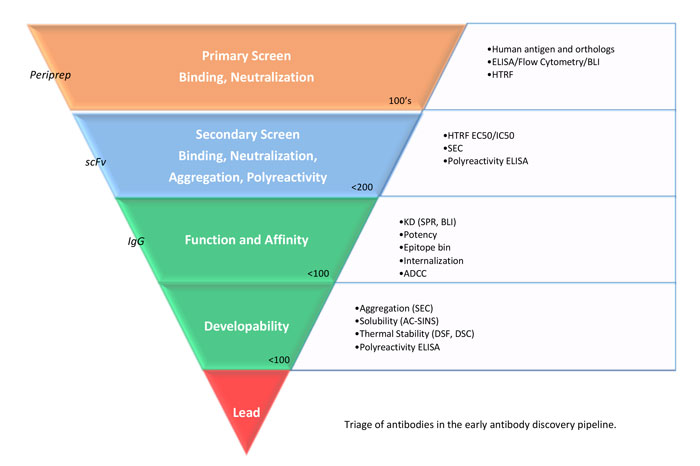

Figure 2: Triage of antibodies in the early antibody discovery pipeline
HTS for Developability
Assessing developability of antibodies is critical in determining a successful path forward for a hit with the desired functional profile.7 SEC may be employed to assess aggregation potential at high concentration formulations required for clinic. Methods such as Differential Scanning Calorimetry (DSF) enable early-stage assessment of thermal stability in an RT-PCR machine, although a simpler approach of ‘cook and bind’ supernatant ELISAs may be utilised.8,9 Antibody specificity reduces potential immunogenicity or off-target effects in the clinic. Therefore, testing cross-reactivity to antigen family members, and non-relevant proteins or cellular preps is important, and correlates with clearance in mice.10-12 Antibody solubility may be impacted by IgG self- or cross-interaction leading also to increased viscosity and aggregation. Affinity capture self-interaction nanospectroscopy (AC-SINS) utilises gold nanoparticles pre-coated with polyclonal capture IgG which immobilises test IgG, and then measures plasmon wavelength between clusters formed by self-interaction.13
This assay has been adapted to high-throughput with supernatants. As it correlates with lower-throughput assays including cross-interaction chromatography, and ultimately in vivo clearance, it is fast becoming essential in many labs. In parallel with experimental data, in silico developability assessment pipelines have been developed, using antibody sequence analysis and machine learning to predict behaviour of molecules, particularly aggregation.14 The main challenge in this area is the complex analysis of the experimental and in silico data – determination of thresholds, prioritisation of parameters, and subsequent in vivo correlation and prediction of pharmacokinetics. Whilst several groups have suggested strategies to overcome this challenge, it may take further assessment of large datasets of antibodies throughout the antibody discovery pipeline to achieve robust criteria for industry.10, 15
The rest of this content is restricted - login or subscribe free to access


Why subscribe? Join our growing community of thousands of industry professionals and gain access to:
- quarterly issues in print and/or digital format
- case studies, whitepapers, webinars and industry-leading content
- breaking news and features
- our extensive online archive of thousands of articles and years of past issues
- ...And it's all free!
Click here to Subscribe today Login here
Related organisations
Abimmune Technologies Inc., Centre for Drug Research and Development Canada