High-throughput screening platforms incorporating physiologically relevant 3-D models
Posted: 8 January 2017 | Anton Simeonov, Madhu Lal-Nag | No comments yet
Two-dimensional monolayer cell proliferation assays for cancer drug discovery have enabled large-scale screens. But they represent a simplified view of oncogenes or tumour suppressor genes as the genetic drivers of cancer cell proliferation.

There is now growing evidence that the cellular and physiological context in which these oncogenic events occur plays a key role in how they drive tumour growth in vivo, and therefore in how tumours respond to drug treatments. In vitro three-dimensional spheroid tumour models are being developed to better mimic the physiology of tumours in vivo in an attempt to improve the predictability and efficiency of drug discovery for the treatment of cancer.
Traditionally, the screening of a large collections of compounds to discover new cancer drugs has been done using cell proliferation assays in which cells grow as monolayers attached to plastic surfaces.1 However, the limitations imposed by these 2-D monolayer cultures have become apparent only recently. Prolonged cell culture of 2-D monolayer cells on plastic surfaces can significantly change their response to anticancer therapeutic agents.2 Examples of this are chemotherapeutic agents such as cisplatin and fluorouracil, which show significant toxicity in 2-D monolayers but very little efficacy in 3-D cultures. On the other hand, certain drugs like trastuzumab show significant activity in 3-D cultures with little to no effect in 2-D monolayers.3 In addition to differences in the effects of drugs, transcriptome analysis of cancer cell lines that represent different tumour types are very different from primary tumours. Furthermore, there is now ample evidence that the tumour microenvironment is critical for tumour physiology and pharmacological responses to drug treatments in vivo.4
Although current monolayer growth assays on plastic capture the proliferative aspect of cancer cells, the cellular complexity of the tumour and the interactions with its microenvironment are not included and, as a consequence, the activity of drugs in these assays can be quite different from their activity in vivo. In vivo, tumour physiology is modulated by its niche through direct cell-to-cell contact secreted signalling molecules and physical properties of the environment like low pH or oxygen levels.5 These factors modify drug penetration into the tumour, regulate expression of cellular drug transporters, modulate signalling pathways, and in general activate mechanisms that in many cases render the cells less susceptible to the effects of drugs.6 It is therefore critical to develop assays in which the tumour microenvironment is recapitulated, at least in part, so that the effects of the drugs better mimic those seen in clinic for native tumours.
Building the right model
The choice of 3-D model and the end point for toxicity/efficacy testing is dependent on both the physiological question to be answered and the scale of the screen.7 Although very powerful and promising, the use and predictivity of 3-D models has to be carefully validated for each given application, and conditions of cultivation and sample collection need to be standardised and controlled. For screening purposes, 3-D cell culture models can be treated with many classes of substances (eg, small molecules, biologicals, siRNA).8 3-D spheroid assays such as the soft agar anchorage-independent growth and clonogenic growth assays have been used for many years, and the challenge for their use in drug discovery is the inability to format them in high throughput screening microtitre plates.9-10
Now, several techniques are available which enable the formation of spheroids in 96 and 384-well microtitre plates. Screening platforms that allow formation of spheroids by enhancing cellular aggregation include round bottom plates with ultralow adherent (ULA) surfaces or the hanging drop techniques.11 Spheroids of sizes ranging from ~50 to 500µm in diameter have been grown in wells of microtitre plates using these techniques. Smaller spheroids measuring up to 200µm may indeed be sufficient to reflect cell-cell and cell-matrix interactions, but most likely do not reflect pathophysiological conditions with hypoxic areas in the spheroid center, and proliferation and nutrient gradients. Hypoxia is not only a well-established direct radio- and drug-resistance factor but it can also lead to numerous indirect effects in tumour cells by modulating gene expression patterns such as that of the HIF1-alpha-regulated genes.12-13
The successful growth of spheroids in a microtitre plate is cell dependent, most likely reflecting expression of cell-cell adhesion proteins, eg, E-cadherin, secretion of extracellular matrix (ECM) products, collagen and fibronectin, and requires the careful testing of media conditions as well as plate types.14 Media that enhance the “stemness” of cells, eg, low serum, and FGF- and EGF-containing media, also help induce formation of spheres.15 In this regard, studies have shown that cells obtained from spheroid cultures are more tumorigenic in vivo, and might be enriched for cancer stem cell populations that are postulated to be those responsible for tumour initiation, metastasis, and recurrence after drug treatment.
Answering biological questions
To answer different biological questions, a range of 3-D models have been established, as shown in Figure 1. In order to study the effect of drug treatment on spheroids that recapitulate certain aspects of a primary tumour, including hypoxia, pH gradients and others, we report the establishment of an HTS-compatible, real-time 3-D spheroid formation assay in 384-well round bottom ULA plates (manuscript accepted for publication). The platform described uses a Hey-A8 cell line constitutively expressing GFP, which enables real-time measurements of spheroid size. Additional endpoint assays, such as ATP levels (CellTiter Glo) to measure cell viability, propidium iodide (PI) to measure cell death, and bright field microscopy to characterise the viability and size of the spheroids, are used to further benchmark pharmacological responses to chemotherapeutic drugs with different mechanisms of action. This recent trend towards multiplexing assay readouts yields the most comprehensive information on a variety of different biological mechanisms. Similarly, as shown in Figure 1, different HTS models need to be developed based on specific biological questions.
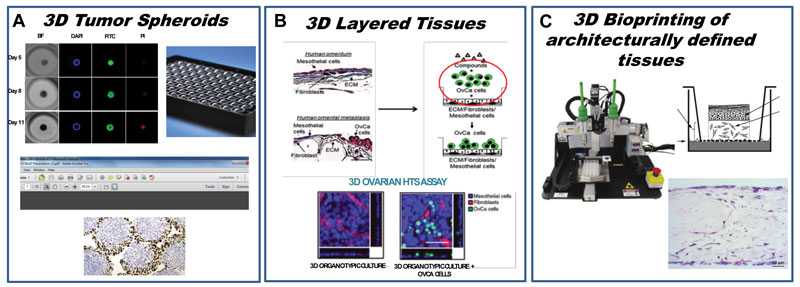
Figure 1. Development of a continuum of platforms of physiologically relevant 3-D HTS assays: (A) Tumor spheroid and organoid HTS; (B) HTS assay to measure inhibition of omental metastasis; (C) Bioprinting of human oncology disease models.
To investigate the effect of small molecules on metastasis in ovarian cancer, we developed an HTS-compatible assay that measured the inhibition of ovarian cancer epithelial cell line Hey A8, tagged with GFP as described above, to migrate through a 3-D-reconstructed omentum comprised of patient fibroblasts and mesothelial cells. This model was built to mimic an in vivo scenario of metastasis.^16 Furthermore, in addition to HTS 3-D miniaturised screening models, bioprinting of human tissue represents a paradigm shift in benchmarking the screening models being developed. As shown in Figure 1, bioprinting human oncology disease models serves as the platform for testing the chemotherapeutic effect of hit compounds chosen from HTS assays. The hope is that this will lead to earlier/cheaper attrition in the number of compounds that will likely fail in animal models and, in turn, serve to iteratively inform better design of HTS 3-D models.
As a rule, treatment with a chemotherapeutic agent can affect cellular and/or 3-D cell culture morphology, viability, metabolic activity or tissue-specific function.5 Here, the repertoire of possible tissue-specific phenotypic end points is constantly widening, together with the development of specialised 3-D tissue models. An example of this can be seen in the case of liver microtissues of co-cultured hepatocytes and non-parenchymal cells (NPCs). Established approaches include monitoring albumin and urea secretion, bile acid secretion, Kupffer cell-dependent IL-6 and TNF-alpha secretion, to name a few.17 Contractile responsiveness of the myocardial microtissue model or glucose-stimulated insulin secretion by pancreatic microislets are newer additions to the growing list of phenotypic endpoints.18 Additionally, cultivated 3-D cell culture models can be further analysed using transcriptomic and proteomic methods, allowing for RNA and protein expression profiling upon treatment with chemotherapeutic agents.
Reproducibility in High-throughput screening
One of the main concerns with any cell-based assay remains reproducibility and robustness from run to run. In 3-D assays, as well as assays that use primary cells, given the complexity of the system being miniaturised, the better part of assay development should concentrate on demonstrating that the assays are indeed robust and reproducible, thereby ensuring that the pharmacological responses are consistent over time. This is illustrated in Figure 2, which delineates the assay development process for an HTS 3-D organoid assay using ovarian cancer cell line, Hey A8-GFP. Assay development to ensure robustness and reproducibility was conducted using a laser scanning cytometer, the Acumen eX3, over the course of eight days to ensure Z’ values over 0.5 over the course of organoid growth, treatment with chemotherapeutics, and assessment of phenotypic response.
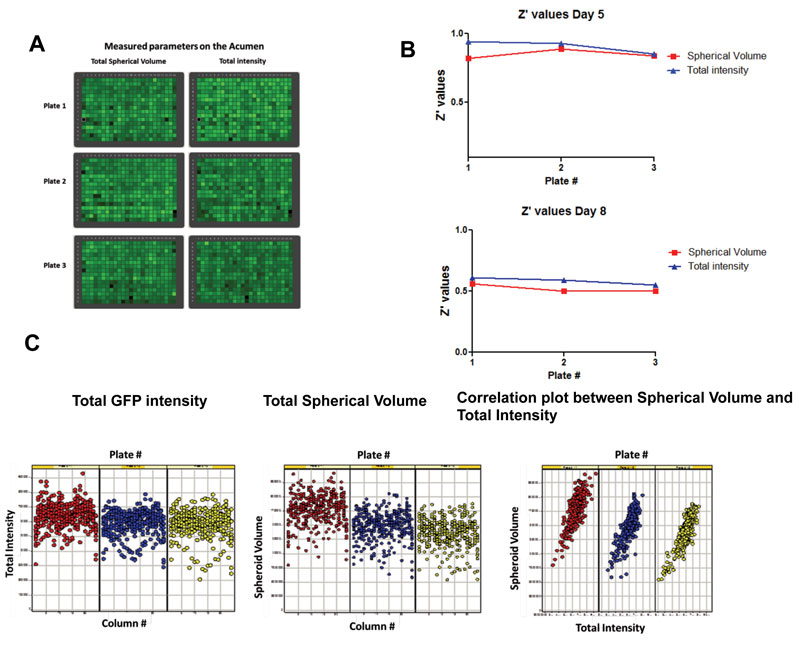
Figure 2. Development of a robust and reproducible HTS organoid assay: (A) Heat map of Acumen measurements for total spherical volume and total intensity; (B) Z’ values for spherical volume and total intensity over the time course of the experiment; (C) Correlation of total GFP intensity and spherical volume across plates and with each other in a single plate indicating robustness and uniformity of signal.
Conclusion
Three-dimensional cultures hold significant promise for the selection of optimal anticancer therapeutic agents and the advancing of candidates to preclinical studies. This will lead to a significant decrease in the attrition of drug failure rates in clinical trials. However, there are many challenges yet to be overcome in the development of prognostic models with respect to efficacy. Currently available high-throughput three-dimensional models tend to recreate only a subset of in vivo conditions. Consequently, the effect of a therapeutic agent on a 3-D model cannot be studied in its entirety as relates to its counterpart in vivo. Furthermore, a gap exists between the biological questions being asked of 3-D in vitro models and the technical capabilities of the high-content and high-throughput systems to answer those questions.
The development of our 3-D screening platform is a step towards understanding the contribution of the tumour microenvironment to cancer cell viability. There is a dire need for chemotherapeutic combinations that will selectively attack and kill both the actively proliferating cancer cells and the slower growing cancer stem cell population that is responsible for generating the drug-resistant cohort of cells that survive and repopulate the tumour. The work being done in this arena aims to study and understand the role of tumour heterogeneity, clonal evolution, dormancy and cell death, and in doing so, to bring forth novel mechanisms of action that will help uncover druggable targets. In the long term, we hope that these HTS 3-D models being developed represent a paradigm shift in the way we view and define relevant biological models and that this screening regimen will be broadly applied to more than one disease type.
About the authors
 ANTON SIMEONOV is the Scientific Director of the Division of Pre-Clinical Innovation (DPI) at the National Center for Advancing Translational Sciences (NCATS), where he oversees the intramural research operations of the Center. He received his PhD in bioorganic chemistry from the University of Southern California and a BA in chemistry from Concordia College, and trained as a postdoctoral fellow at the Scripps Research Institute, La Jolla, CA. Dr. Simeonov’s current research interests include novel detection chemistries and techniques, assays and devices for diagnostics, assay miniaturisation, and novel approaches to screening and therapeutics development.
ANTON SIMEONOV is the Scientific Director of the Division of Pre-Clinical Innovation (DPI) at the National Center for Advancing Translational Sciences (NCATS), where he oversees the intramural research operations of the Center. He received his PhD in bioorganic chemistry from the University of Southern California and a BA in chemistry from Concordia College, and trained as a postdoctoral fellow at the Scripps Research Institute, La Jolla, CA. Dr. Simeonov’s current research interests include novel detection chemistries and techniques, assays and devices for diagnostics, assay miniaturisation, and novel approaches to screening and therapeutics development.
 MADHU LAL-NAG currently serves as the head of the Trans NIH RNAi Facility at the National Center for Advancing Translational Sciences (NCATS), which is responsible for developing and conducting genome-wide physiologically relevant phenotypic assays for Intramural researchers. Madhu joined NCATS in 2013, where she worked as a research scientist primarily to develop an assay platform of 3-Dimensional physiologically relevant, multi-cell-type disease models that are amenable to high-throughput screening. Prior to joining NCATS, she completed her postdoctoral fellowship at the National Institute on Aging.
MADHU LAL-NAG currently serves as the head of the Trans NIH RNAi Facility at the National Center for Advancing Translational Sciences (NCATS), which is responsible for developing and conducting genome-wide physiologically relevant phenotypic assays for Intramural researchers. Madhu joined NCATS in 2013, where she worked as a research scientist primarily to develop an assay platform of 3-Dimensional physiologically relevant, multi-cell-type disease models that are amenable to high-throughput screening. Prior to joining NCATS, she completed her postdoctoral fellowship at the National Institute on Aging.
Acknowledgements
We would like to acknowledge Dr Marc Ferrer, Dr Jiaqi Fu, Daniel Fernandez, and Steve Titus, for their expertise in the conception, experimental execution and imaging of our HTS and high-content platform for 3-D assays. We would also like to acknowledge the Trans NIH RNAi Facility for supporting this research.
References
- Breslin S, O’Driscoll L. Three-dimensional cell culture: the missing link in drug discovery. Drug Discov Today. 2013;18(5-6), 240-9
- Kim JB. Three-dimensional tissue culture models in cancer biology. Semin Cancer Biol. 2005;15 (5): 365-77
- Das V, Bruzzese F, Konecny P, Iannelli F, Budillon A, Hajduch M. Pathophysiologically relevant in vitro tumor models for drug screening. Drug Discov Today. 2015;20(7): 848-55
- McMillin DW, Negri JM, Mitsiades CS. The role of tumour-stromal interactions in modifying drug response: challenges and opportunities. Nat Rev Drug Discov. 2013;12(3): 217-28
- Kunz-Schughart LA. Multicellular tumor spheroids: intermediates between monolayer culture and in vivo tumor. Cell Biol Int. 1999;23(3): 157-61
- Mueller-Klieser WF, Sutherland RM. Oxygen tensions in multicell spheroids of two cell lines. Br J Cancer. 1982;45(2): 256-64
- Haas J, Manro J, Shannon H, et al. In Vivo Assay Guidelines. In Assay Guidance Manual. Sittampalam GS, Coussens NP, Brimacombe K, Grossman A, et al. Eds. Eli Lilly & Company and the National Center for Advancing Translational Sciences: Bethesda (MD), 2004. Most recently accessed on 15/01/2017
- Balis FM. Evolution of anticancer drug discovery and the role of cell-based screening. J Natl Cancer Inst. 2002;94(2): 78-9
- Tung YC, Hsiao AY, Allen SG, Torisawa YS, Ho M, Takayama S. High-throughput 3D spheroid culture and drug testing using a 384 hanging drop array. Analyst. 2011;136(3): 473-8
- Tanner K, Gottesman MM. Beyond 3D culture models of cancer. Science translational medicine. 2015;7(283): 283ps9.
- Tannock IF, Lee CM, Tunggal JK, Cowan DS, Egorin MJ. Limited penetration of anticancer drugs through tumor tissue: a potential cause of resistance of solid tumors to chemotherapy. Clin Cancer Res. 2002;8(3): 878-84
- Zanoni M, Piccinini F, Arienti C, et al. 3D tumor spheroid models for in vitro therapeutic screening: a systematic approach to enhance the biological relevance of data obtained. Scientific report. 2016;6: 19103
- Trask OJ, Jr. Nuclear Factor Kappa B (NF-kappaB) Translocation Assay Development and Validation for High Content Screening. In Assay Guidance Manual. Sittampalam GS, Coussens NP, Brimacombe K, et al. Eds. Eli Lilly & Company and the National Center for Advancing Translational Sciences: Bethesda (MD). 2004
- Zschenker O, Streichert T, Hehlgans S, Cordes N. Genome-wide gene expression analysis in cancer cells reveals 3D growth to affect ECM and processes associated with cell adhesion but not DNA repair. PLoS One. 2012;7(4): e34279.
- Schiavoni G, Gabriele L, Mattei F. The tumor microenvironment: a pitch for multiple players. Front Oncol. 2013;3: 90
- Kenny HA, Lal-Nag M, White EA, et al. Quantitative high throughput screening using a primary human three-dimensional organotypic culture predicts in vivo efficacy. Nature communications. 2015;6: 6220
- Martin S, Buehler G, Ang KL, et al. Cell-Based RNAi Assay Development for HTS. In Assay Guidance Manual. Sittampalam GS, Coussens NP, Brimacombe K, et al. Eds. Eli Lilly & Company and the National Center for Advancing Translational Sciences: Bethesda (MD). 2004
- Riss TL, Moravec RA, Niles AL, et al. Cell Viability Assays. In Assay Guidance Manual, Sittampalam GS, Coussens NP, Brimacombe K, et al. Eds. Eli Lilly & Company and the National Center for Advancing Translational Sciences: Bethesda (MD). 2004
Related topics
Assays, Cell Cultures, High-Throughput Screening (HTS), Molecular Modelling