High-content imaging: challenges of the 3D world
Posted: 17 June 2019 | Jeremy C. Simpson, Margaritha M. Mysior | No comments yet
The potential of three-dimensional cell culture models is now well-recognised by scientists; however, there are still some challenges. Margaritha Mysior and Jeremy Simpson address some of the common challenges of working with spheroids and organoid models – including how they are developed and how to obtain high-resolution, high-content images and how to develop three-dimensional high-resolution imaging assays.


ALTHOUGH the famous phrase “a picture paints a thousand words”, attributed to the marketing executive Frederick R Barnard, first entered common use in the English language almost 100 years ago, its origins are thought to date back several centuries. However, this old adage has never been more relevant than now in the twenty-first century. Fluorescence microscopy images and their exploitation to understand cell function is arguably the most fundamental resource employed today in research laboratories around the world. Their value lies in the fact that visualisation of molecules, organelles and their interactions and behaviour is now easily facilitated through an ever-expanding palette of fluorescence reporters and tools. These can be applied in real time and in living material, giving researchers unprecedented access to the inner workings of cells.
However, the paradigm shift in recent years has been the transition to quantitative imaging, such that now the most complex of phenotypes observed in an image, or indeed in some cases not even visible to the human eye, can be described in mathematical ways. This evolution of quantitative microscopy is referred to as high-content imaging, emphasising that the image itself is a rich source of information. In turn, the quantitative interpretation of images is achieved through high-content analysis (HCA).1 The power of this technology is that it can be applied in a highly reproducible and high-throughput manner allowing researchers to delineate cellular pathways on a systems-wide scale, thereby enabling drug discovery and development in the global pharmaceutical industry.2
To facilitate the HCA process, the vast majority of experiments reported to date have utilised cultured cells effectively growing in two dimensions in flat, optically-transparent multi-well plates. This format is convenient from a cell culture perspective and presents relatively few problems for advanced automated microscopes to gather millions of in-focus images in short periods of time (Figure 1). However, while such experiments are certainly informative, this format does not accurately represent the arrangement of cells in complex tissues and organs, and therefore has limitations.
One can envisage that events such as cell-cell signalling and communication – the interplay between the different cell types found in tissues and the mechanisms by which therapeutics transit across cellular layers – are not necessarily reliably recapitulated from simple cell monolayer models. It is therefore clear that the next phase in our deeper understanding of such events in cells will only be realised through models that more closely resemble the in vivo situation.


Fully-automated high-content screening microscope
Methods of producing three-dimensional cell models
Although three-dimensional cell culture models have been described for several decades, it is only in the last few years that their potential has been more widely appreciated by biologists.3 This is largely a result of new technical developments that facilitate easier production in the laboratory, coupled with a new generation of automated confocal microscopes that can rapidly capture information from multiple planes within them. Most multicellular three-dimensional models take the form of spheroids, which can be of a solid cell mass or hollow core format, or organoids, which are morphologically similar to spheroids but derived from multiple cell types. Most commonly, spheroids and organoids can be assembled in one of four ways: 1) simple aggregation in U-bottomed wells, 2) aggregation in a liquid droplet, 3) self-directed assembly in, or on, a matrix or scaffold, and 4) in a microfluidic or micropatterned device. Each of these methods has its respective advantages and disadvantages, typically linked to the level of control of spheroid/organoid assembly and scale of their production. From a fluorescence imaging perspective, the aggregation methods in plates or droplets have the advantage that typically a single spheroid per well is formed, meaning that its identification in the imaging process, assuming that it is centrally located, is relatively straightforward. However, this approach is generally considered as low-throughput and is not easily scalable for high-throughput imaging-based screens. Matrix- and scaffold-based methods allow for much higher numbers of spheroids to be produced per well, typically upwards of 500 assemblies per well in a 96-well plate, for example; although this approach suffers from problems associated with autofluorescence from the matrix and the fact that spheroids assemble randomly across the well, including at multiple heights. This means that large numbers of confocal image slices may be required to capture the full depth of the spheroid, resulting in lengthy image acquisition times. Microfluidic and micropatterned devices are attractive for spheroid generation as they can be custom-designed and provide a highly controlled environment, but they can be time-consuming and technically challenging to manufacture, and/or expensive to purchase on a large scale. Regardless of the method of spheroid generation selected, they present several opportunities for the next generation of cell-based assays that need to consider the local microenvironment. The challenge, therefore, is how to both transition well-established assays in two-dimensional form into these cell models, as well as develop novel assays with better tissue and/or clinical relevance.
The challenges of performing assays in three-dimensional models
There are several obstacles to establishing assays in three-dimensional models. Many of these relate to the heterogeneity of spheroids within any given population, with respect to the cell numbers within each spheroid, as well as their shape, size and distribution within the structure. In addition, many cell-based assays involve multiple manipulation steps, which can add complexity and unreliability to the assay. Also of note is that manipulations and perturbations applied to the cells (for example, transfections and drug treatments), which are highly routine in two-dimensional models, may not be equal across all cells within a spheroid. From an imaging perspective, consistent staining of structures of interest is essential to avoid quantification errors between cells within the same spheroid. Consideration should also be given to the fact that resolution is typically three-fold inferior in the z-dimension, compared to that in the xy plane. Furthermore, optical aberrations can become significantly deep inside spheroids, particularly in structures greater than 150μm in diameter. Some of these issues can be resolved by greater standardisation of spheroid assembly, particularly using production methods that offer a higher degree of control. Standardisation of spheroid size and shape is undoubtedly of critical importance in drug development and delivery screens; with, for example, the efficacy of penetration of therapeutics across multicellular layers understood to be directly influenced by the arrangement of the cells. While not suitable for all applications, micropatterning of spheroids in highly regular arrays offers exciting possibilities for assay standardisation in drug delivery applications, as it allows highly uniform production of spheroids of defined size and potentially shape (Figure 2).
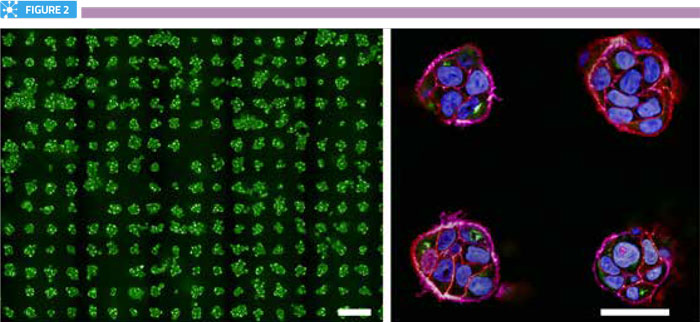

Arrays of spheroids. Left panel shows a portion of a single well from a micropatterned 96-well plate containing over 250 mini-spheroids. Scale bar is 100μm. Right panel shows zoomed view of four spheroids, labelled for nuclei (blue), actin (purple), cell-cell junctions (red) and the Golgi complex (green). Scale bar is 40μm.
Data, data, and still more data
Once cell-based assays and appropriate imaging regimes have been established in a three-dimensional format, the next challenge to be addressed is how to extract meaningful data from the images. Use of HCA algorithms to precisely segment individual cells and subcellular structures from monolayer-grown cells is now routine, and quantification is highly reliable. The situation in spheroids, however, is more complex, as individual subcellular objects of interest must first be associated with the correct cell, and then each cell with the spheroid; and all of this with consideration to the three-dimensional space. To be successful, it is therefore essential that a sufficiently high image-sampling frequency is applied in the z-plane through the complete volume of the spheroid. If this is achieved, HCA potentially has the power to provide rich quantitative information describing subcellular morphologies and phenotypes as well as, for example, the efficacy of penetrance of therapeutics to particular cells within the different layers of the spheroid or organoid. Such information is undoubtedly of huge potential benefit to the drug discovery community. While there is excitement about the knowledge that such experiments can deliver, this deep level of content is extremely challenging in terms of data handling. For example, imaging of approximately 750 small four-day-old spheroids, with a diameter of approximately 60μm, in a single well of a 96-well plate requires the acquisition of at least 50 optical slices in the z-direction in order to capture the complete volume of the objects. For a dual colour channel image this equates to 45Gb of data per well and 4.3Tb of data per plate. Larger spheroids have significantly greater depth, requiring many more z-planes to be acquired, and the data volume can easily approach 12Tb for an entire 96-well plate. Clearly, such massive datasets not only present problems from the perspective of image analysis, but there are implications for the feasibility of how such data volumes can be stored and mined.
Three-dimensional spheroid and organoid models present an exciting opportunity to bridge the knowledge gap between our understanding of cell function obtained from classical monolayer cultures and that coming from tissues. This gap is arguably of critical importance in the field of drug discovery and delivery, with still too many pharmaceuticals failing at late stages in the development pipeline process. Although there are clearly several challenges associated with transitioning fluorescence assays into three-dimensional models, they nevertheless offer a new avenue to be explored, and the pharmaceutical industry in particular would be foolish to ignore their potential. Perhaps in the not too distant future, even ten thousand words may be insufficient to describe the information that spheroid images (or pictures) hold.
Biographies




References
- Meijering E, et al. (2016) Imagining the future of bioimage analysis. Nature Biotechnology 34: 1250-1255.
- Fraietta I, Gasparri F. (2016) The development of high-content screening (HCS) technology and its importance to drug discovery. Expert Opinion on Drug Discovery 11: 501-514.
- Simian M, Bissell MJ. (2016) Organoids: a historical perspective of thinking in three dimensions. Journal of Cell Biology 216: 31-40.
Related topics
Assays, Big Data, Drug Delivery, Imaging
Related organisations
University College Dublin
Related people
Jeremy C. Simpson, Margaritha M. Mysior