Gene map identifies genes unique to different cancers
Posted: 26 November 2015 | Victoria White | 2 comments
Scientists from the University of Toronto have switched off, one by one, almost 18,000 genes – 90% of the entire human genome – to find the genes that are essential for cell survival…
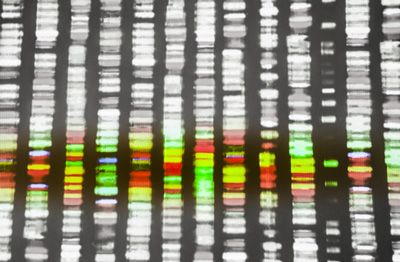

Scientists from the University of Toronto have switched off, one by one, almost 18,000 genes – 90% of the entire human genome – to find the genes that are essential for cell survival.
The data revealed a “core” set of more than 1,500 essential genes.
By turning genes off in five different cancer cell lines, including brain, retinal, ovarian, and two kinds of colorectal cancer cells, the team uncovered that each tumour relies on a unique set of genes that can be targeted by specific drugs. The finding raises hope of devising new treatments that would target only cancer cells, leaving the surrounding healthy tissue unharmed.
“It’s when you get outside the core set of essential genes, that it starts to get interesting in terms of how to target particular genes in different cancers and other disease states,” says Professor Jason Moffat, from the University of Toronto’s Donnelly Centre.
Because different cancers have different mutations, they tend to rely on different sets of genes to survive. Moffat’s team have identified distinct sets of “smoking gun” genes for each of the tested cancers – each set susceptible to different drugs.
“We can now interrogate our genome at unprecedented resolution in human cells that we grow in the lab with incredible speed and accuracy. In short order, this will lead to a functional map of cancer that will link drug targets to DNA sequence variation,” says Moffat.
Data can point to more precise treatments for different cancers
His team has already shown how this can work. In his study, metformin, a widely prescribed diabetes drugs successfully killed brain cancer cells and those of one form of colorectal cancer – but was useless against the other cancers he studied. However, the antibiotics chloramphenicol and linezolid were effective against another form of colorectal cancer, and not against brain or other cancers studied. These data illustrate the clinical potential of the data in pointing to more precise treatments for the different cancers.
“The Moffat group has developed a powerful CRISPR library that could be used by investigators around the world to identify new treatment strategies for the treatment of cancer,” says Aaron Schimmer, a professor in the Department of Medical Biophysics and a medical oncologist at Princess Margaret Cancer Centre, who was not involved in the study. “I would be interested in using this tool to identify new treatment approaches for acute myeloid leukemia – a blood cancer with a high mortality rate.”
Related topics
Antibiotics, Gene Testing
Related conditions
Diabetes
Related organisations
Toronto University





I am ready to cooperation for joint researches. The insufficient understanding of the mechanism of oxygen transport in the conditions of formation of cancer tumors follows from your data.
The Oxygen generation.
Process of oxygen transport provides generation of Oxygen in structure of DNA. The sequence of an empirical component of a genome contains the incomplete (open) membrane volume of an oxygen exchange. These are various phases of empirical symbols are carriers of various nucleon functions in structure of neutrons of Oxygen. In this case the functional model of structure of all isotopes of Oxygen is used. The main genetic feature of this structure is the mechanism of functional reorientation of values of transport of a phase exchange.
This mechanism contains superthin structures of functional mutations. Definition and development of these processes contain and in neural structures of a brain.
In case of your interest I can provide more detailed research material.