Computational epitope map of SARS-CoV-2 Spike protein developed
Posted: 7 May 2021 | Victoria Rees (Drug Target Review) | 1 comment
Researchers have produced a computational simulation of the SARS-CoV-2 Spike protein, finding spots that glycans do not cover.
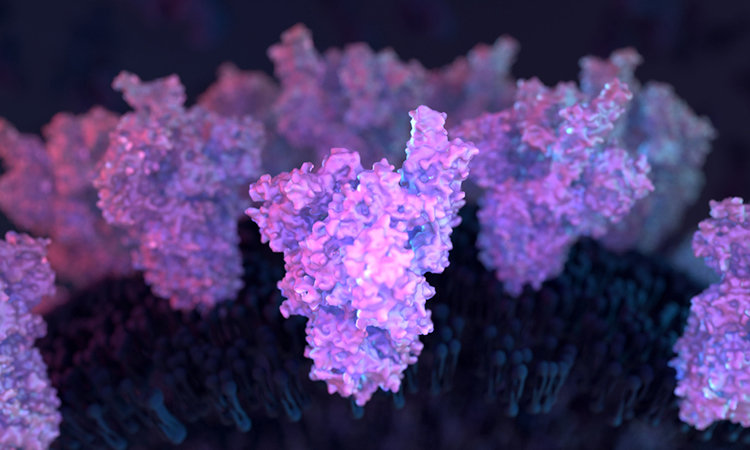

Researchers have produced a dynamic model of the SARS-CoV-2 Spike (S) protein, revealing potential drug targets to treat COVID-19. The study was conducted at the Max Planck Institute of Biophysics, Germany.
According to the team, the portrait was created via computer modelling from sophisticated three-dimensional (3D) simulations of the S protein in action. The researchers used a computer application called molecular dynamics (MD) simulation to model the conformational changes in the S protein on a time scale of a few microseconds. The aim was to map its many shape-shifting manoeuvres accurately at the atomic level in hopes of detecting exploitable structural vulnerabilities to combat the virus.
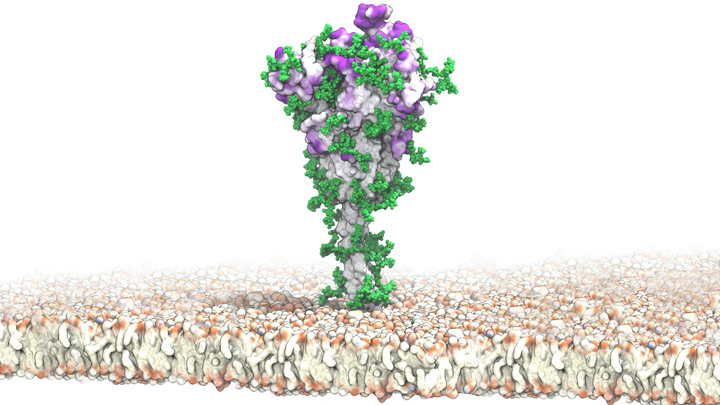

SARS-CoV-2 S protein [credit: Sikora M, PLoS Comput Biol, 2021].
The researchers highlight the many chain-like structures (green) that adorn the protein’s surface (white). These are sugar molecules called glycans that are thought to shield the S protein by sweeping away antibodies. Other areas (purple) that the simulation identified are the most-attractive targets for antibodies, based on their apparent lack of protection by those glycans.
The simulations suggest that glycans act as a dynamic shield on the S protein. Rather than being fixed in space, those glycans sweep back and forth to protect more of the protein surface than initially meets the eye.
However, the team emphasise that the glycans miss spots of the protein just beyond their reach. It is those spots that the researchers suggest might be prime targets on the S protein that are especially promising for the design of future vaccines and therapeutic antibodies.
This same approach can now be applied to identifying weak spots in the coronavirus’s armour. It may also help researchers understand more fully the implications of newly emerging SARS-CoV-2 variants. The hope is that by capturing this virus and its most critical proteins in action, scientists can continue to develop and improve upon vaccines and therapeutics.
The study was published in PLoS Computational Biology.
Related topics
Disease Research, Informatics, Molecular Targets, Protein, Proteomics, Target Validation
Related conditions
Covid-19
Related organisations
Max Planck Institute of Biophysics






Hi,
Excellent work!!!
1) does the algorithm offer insights as to the role of s100 proteins in the pathogensis of COVID-19?
2) Does it allow to understand the contribution of spike/non spike viral proteins in scaffolding transformational changes during infection/attachment that may give rise to new variants as well as host conformational changes?
3) Any indication of the impact of host surface receptors and virus spike proteins on infectivity?
Thank you