The science behind the systematic discovery of molecular glues
Posted: 25 September 2025 | Drug Target Review | No comments yet
For decades, molecular glues have been stumbled upon rather than designed. A new scientific approach is now changing that – expanding what is considered druggable.


Drug discovery has long faced the limitations of what is considered ‘druggable’. For decades, the field has focused on proteins with convenient binding pockets that small molecules can latch onto. Yet large areas of the proteome resist such neat classification. The emergence of targeted protein degradation (TPD) has begun to address these limitations, with GlueSEEKER® offering a structured way to explore molecular glue mechanisms.
We sat down with Dr Jia Lu, Principal Scientist at PhoreMost and co-developer of GlueSEEKER, to discuss the challenges of molecular glue discovery, the promise of systematically reprogramming E3 ligases and what lies ahead for this rapidly advancing area of research.
From Cambridge to PhoreMost
Jia’s path into drug discovery began with a PhD in pathology at the University of Cambridge, followed by postdoctoral research at the Babraham Institute. “I joined PhoreMost in 2021 and since then, have been working on projects related to TPD, focusing on target discovery and target identification using our proprietary SITESEEKER technology,” he explains.
That work naturally led her to the cutting edge of molecular glue research. Most recently, Jia co-developed GlueSEEKER, a platform designed to transform how molecular glues are discovered and deployed. “The platform is designed to address the challenges associated with the systematic discovery of molecular glues,” he says.
Why molecular glues are so hard to find
Molecular glues are not conventional small molecules; they do not simply block or activate proteins by binding to them. Instead, they induce or enhance protein–protein interactions (PPIs), effectively ‘gluing’ a target protein to an effector such as an E3 ligase, which can then trigger degradation.
But therein lies the problem: these are three-way interactions. As Jia points out, “The tri-partite nature of the PPIs means canonical screening tools, which focus on binary interactions, are not suitable for novel molecular glue discovery.”
Due to the lack of knowledge surrounding structural compatibility at the PPI interface, the discovery of targeted E3 ligase:protein of interest (POI) interactions carries high risk.
In practice, this means most of the standard tools used in drug discovery are simply not equipped to deal with the complexity of molecular glues. They can measure whether two molecules interact, but not whether a third can come in to stabilise or redirect that interaction. As a result, whole classes of potential drug mechanisms have remained poorly understood.
Traditional discovery has leaned heavily on luck. “Due to the lack of knowledge surrounding structural compatibility at the PPI interface, the discovery of targeted E3 ligase:protein of interest (POI) interactions carries high risk,” Jia explains. “As a result, mechanisms-of-action are often only determined post-discovery, rendering their identification largely serendipitous.”
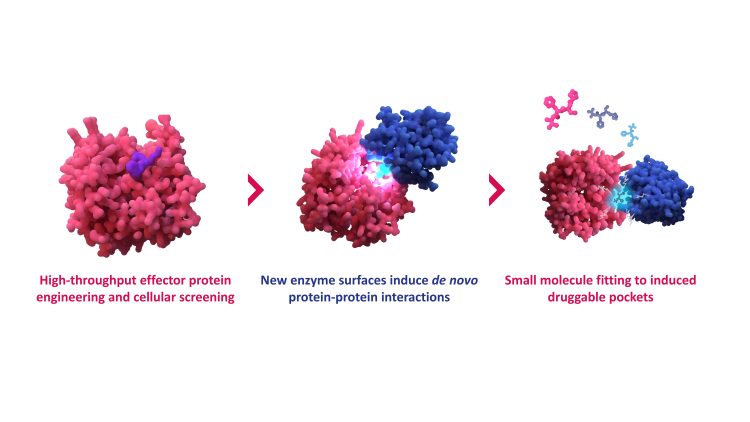

Overview of the GlueSEEKER technology, blending high-throughput biological data with computational drug discovery approaches to unlock previously undruggable targets.
Discovery made systematic
So how does GlueSEEKER actually work? The platform identifies the very interactions molecular glues induce. Jia outlines the principle: “GlueSEEKER allows molecular glues to be discovered by identifying the PPIs they induce. The platform applies high-throughput screening of engineered effector protein surfaces, in this case E3 ligases, to identify those that confer neomorphic (gain-of-function) activities.”
In this context, gain-of-function means that engineered E3 ligases acquire a new activity that is not present in their natural state – for example, the ability to engage with a different substrate protein and direct it for degradation.
By systematically comparing unmodified and mutant versions of E3 ligases, GlueSEEKER uncovers structural blueprints for rationally designing small molecules. “We can rationally design small molecules that mimic these gain-of-function interactions, effectively guiding the targeted degradation of desired protein substrates,” Jia explains.
Unlike other approaches that rely heavily on pre-existing knowledge of ligase or substrate structures, GlueSEEKER provides a more direct route: systematic, high-throughput screening of PPIs in a physiologically relevant context. It means researchers can explore novel ligase–substrate combinations without needing years of accumulated prior knowledge.
Why reprogramming E3 ligases matters
The scale of this breakthrough becomes clear when considering the druggable universe. Currently, much of it remains inaccessible. “Many clinically important targets remain ‘undruggable’ since they lack small molecule binding pockets, which are traditionally required for drugs to bind in order to inhibit activity, stabilise a conformation or modulate function,” Jia notes.
Molecular glues overcome that limitation by binding the E3 ligase and inducing it to engage new substrates. “When applied systematically and at scale, this principle allows the druggable space to be dramatically expanded even with no prior knowledge of ligands,” he says.
This could mean thousands of previously ‘undruggable’ proteins are suddenly within reach. This is not just theoretical; high-throughput approaches are already accelerating the pace of discovery far beyond what chance findings could deliver.
Case study: proof of principle
A recent PhoreMost paper included a case study demonstrating the platform’s potential. Jia summarises: “The case study adds to the growing recognition of genetic and chemical convergence between neomorphic E3 ligase mutations and molecular glues. It shows that despite relatively little structural information on E3 ligase:POI interactions, novel molecular glue degraders can still be discovered systematically.”
Perhaps most importantly, the approach is transferable. “The consistent structure–function relationship is maintained throughout the pipeline, indicating a logical, rational approach that can be applied to E3 ligases beyond CRBN and those with existing molecular glues, further expanding the druggable space and addressing unmet clinical needs.”
The role of AI and computational tools
With high-throughput screening generating mountains of data, computational tools are not optional – they are essential. “AI and computational tools are important components of the GlueSEEKER platform,” Jia says.
GlueSEEKER is designed to utilise screening outputs to inform subsequent peptide design, improving the efficiency and capacity of future screening efforts.
Their roles are multilayered, predicting interfaces, designing neomorphic mutations and guiding small molecule development. But Jia stresses that GlueSEEKER is not only using computational tools passively; it is also designed to refine them. “GlueSEEKER is designed to utilise screening outputs to inform subsequent peptide design, improving the efficiency and capacity of future screening efforts.”
In practice, this means the platform can generate data that strengthens the models, allowing subsequent experiments to be carried out with greater focus and precision.
Beyond CRBN: what comes next
Cereblon (CRBN) is one of the best-known E3 ligases in molecular glue research. It came to prominence because drugs such as thalidomide derivatives work by binding to CRBN and redirecting it to degrade disease-relevant proteins. This has made it something of a poster child for the field. But PhoreMost is looking much further.
“In our latest GlueSEEKER manuscript, we demonstrated that gain-of-function mutations in CRBN can induce de novo interaction with GSPT1, a cell cycle protein, and can be mimicked by small molecules,” Jia notes.
The real excitement comes from the suggestion that this is not unique to CRBN or even the better-studied ligases. “Our latest results suggest the commonly utilised E3 ligases, CRBN and KBTBD4, are not the only members of this emerging phenomenon,” he says.
The team is also exploring how their mini-protein engineering expertise and high-throughput phenotypic screening can feed richer datasets into AI tools, building a stronger foundation for computational design in the future.
Discovery by design
The striking feature of GlueSEEKER is not just the potential to find new glues – but the shift from discovery by chance to discovery by design. Drug discovery has always been a blend of art and science, but this platform aims to bring order to what has historically been a game of luck.
The implications stretch far beyond oncology or immunology; any disease area with undruggable proteins is now a candidate.
Meet the expert


Related topics
Analysis, Artificial Intelligence, Assays, Cancer research, Computational techniques, Drug Development, Drug Discovery Processes, Drug Targets, Oncology, Small Molecules
Related organisations
PhoreMost
Related people
Dr Jia Lu (Principal Scientist at PhoreMost)