High-res 3D structure of proteins inside living eukaryotic cells determined
Posted: 3 June 2019 | Drug Target Review | No comments yet
Nuclear magnetic resonance measurement and state-of-the-art computational science reveal protein structures in higher eukaryotic cells.
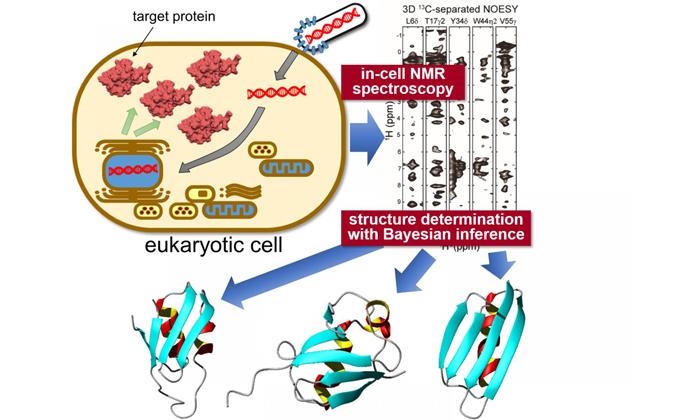

Target proteins are expressed inside sf9 cells, then measured using "in-cell" NMR spectroscopy. Unique statistical analysis with the help of Bayesian inference is applied to calculate the accurate 3D structure of the proteins with unprecedented precision (credit: Tokyo Metropolitan University).
Researchers from Tokyo Metropolitan University have combined ‘in-cell’ nuclear magnetic resonance (NMR) spectroscopy, a bioreactor system and cutting-edge computational algorithms to determine protein structures in crowded intracellular environments for the first time.
The technique promises insight into the intracellular behaviour of disease-causing proteins and novel drug screening applications, allowing in-situ visualisation of how proteins respond to biochemical stimuli.
To tackle the complex and varied internal structure of Eukaryotic cells, the researchers (led by Assistant Professor Teppei Ikeya and Professor Yutaka Ito) applied NMR spectroscopy measurements to specific proteins expressed inside sf9 cultured insect cells.
Biomarkers aren’t just supporting drug discovery – they’re driving it
FREE market report
From smarter trials to faster insights, this report unpacks the science, strategy and real-world impact behind the next generation of precision therapies.
What you’ll unlock:
- How biomarkers are guiding dose selection and early efficacy decisions in complex trials
- Why multi-omics, liquid biopsy and digital tools are redefining the discovery process
- What makes lab data regulatory-ready and why alignment matters from day one
Explore how biomarkers are shaping early drug development
Access the full report – it’s free!
To apply the team’s already-established NMR work to proteins in sf9 cells they combined a sparse sampling-based rapid NMR measurement scheme with state-of-the-art computational methods employing statistical techniques like Bayesian inference – methods tailored to elucidate protein structures efficiently based on a limited amount of structural information from in-cell NMR spectra with inherently low-sensitivity.
A bioreactor system was also equipped inside the NMR apparatus which kept the cells in a healthy state during the measurements.
With this new data, the team were able to elucidate the 3D structure of three model proteins with unprecedentedly high resolution, with a precision of 0.5 Angstroms (0.05 nanometers) for the position of the protein’s main chain atoms. In particular, they identified a significantly different conformation in a localised region of one of the proteins compared to its reference structure in dilute solution.
The conformational difference between proteins ‘in cells’ and ‘in test tubes’ was presumably caused by non-specific interactions with other molecules inside the cells. It is becoming clear that these interactions contribute to the proteins’ biological functions: the ability to locate and quantify structural changes of proteins in an intracellular environment is expected to have a significant impact on biomedical research, making it possible to see how different conditions affect protein conformations in-situ, and quantitatively gauge how treatments impact structural anomalies.
The study has been published online in the journal Angewandte Chemie International Edition.
Related topics
Analysis, Cell Cultures, Imaging, Informatics, Protein, Research & Development, Technology
Related organisations
Tokyo Metropolitan University
Related people
Assistant Professor Teppei Ikeya, Professor Yutaka Ito