Bacteriophage genes could lead to development of new antibiotics
Posted: 8 February 2021 | Victoria Rees (Drug Target Review) | No comments yet
New research has shown that ‘hidden’ lysis genes in bacteriophages could be used in the development of a new class of antibiotics.
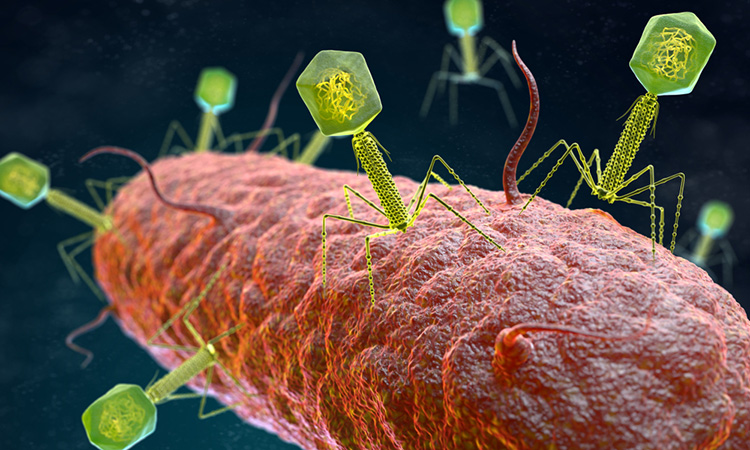

A study has shown how the ‘hidden’ genes in bacteriophages – types of viruses that infect and destroy bacteria – may be key to the development of a new class of antibiotics. The study was conducted at the Center for Phage Technology, part of Texas A&M University’s College of Agriculture and Life Sciences, US.
“There has been an increased interest in bacteriophages and their potential as antibacterial agents to fight pathogenic bacteria,” said Dr Ryland Young, who supervised the study research. “This is in large part due to the ability of the ‘lysis genes’ of the phage to cause a cellular breakdown in the bacterial host.”
According to the researchers, most phages can cause their host cell to rupture, in a process called lysis. They release new ‘progeny’ phage virions that are genetically and structurally identical to the parent virus.
Reduce preclinical failures with smarter off-target profiling
24 September 2025 | 15:00PM BST | FREE Webinar
Join this webinar to hear from Dr Emilie Desfosses as she shares insights into how in vitro and in silico methods can support more informed, human-relevant safety decisions -especially as ethical and regulatory changes continue to reshape preclinical research.
What you’ll learn:
- Approaches for prioritizing follow-up studies and refining risk mitigation strategies
- How to interpret hit profiles from binding and functional assays
- Strategies for identifying organ systems at risk based on target activity modulation
- How to use visualization tools to assess safety margins and compare compound profiles
Register Now – It’s Free!
“Small phages, such as the ones this study focuses on, make a single protein which causes host lysis,” Young said. “The virus produces a ‘protein antibiotic’ that causes lysis in the same way antibiotics like penicillin do – by disrupting the multistage process of cell wall biosynthesis. When the infected cell tries to divide, it blows up because it cannot create the new cell wall between the daughter cells.”
The study focused on characterising the lysis genes of leviviruses, bacteriophages containing small single-stranded RNA genomes with only three to four genes. Among the known levivirus genes is single gene lysis (Sgl). Sgl encodes a protein that induces the cellular breakdown of bacteria.
Many leviviruses contain Sgl genes, but these have remained hidden from researchers as they are small, extremely varied and can be embedded within other genes.
The researchers were able to identify 35 unique Sgls that produced a lytic or destructive effect on E. coli bacteria. They also determined that each of these Sgls could potentially represent a distinct mechanism for the lysis of host cells.
“High mutation rates allow these phages to infect new species of bacteria,” said Dr Karthik Chamakura, the study’s first author. “In order to escape the new hosts, the phages have to either change the existing Sgl gene or evolve a new Sgl. In spite of a very short total length of genomic RNA, these phages can encode two or more Sgls or proto-Sgls for the lytic activity to destroy multiple bacterial hosts.”
The team also observed that a large proportion of the Sgls found in the investigation had originated and evolved within the gene for the phage replication protein (Rep).
“There were a disproportionate number – 22 of the 35 of Sgls or Sgl candidates – found embedded within the Rep gene,” Chamakura said. “Overlaying the location of Sgl genes on the respective Rep sequences revealed that most of the Sgl genes evolved in less conserved regions of Rep. This could mean more highly divergent regions of the levivirus genome, such as the Rep gene, may serve as ‘hotspots’ for Sgl evolution.”
He said the study’s examination of genomes also revealed that closely related phages showed significant evidence of the de novo gene evolution.
“A single-stranded RNA phage might have two or more lysis genes at different stages of gene evolution,” Chamakura said. He concluded that Sgls are extremely diverse and remain vastly untapped as a source for peptides that could be used in protein antibiotics to attack the cellular function of bacteria. He said the study should also be useful in helping uncover small genes and their biological functions in RNA viruses of more complex organisms – such as plants and animals – as well as provide a good model for studying how new genes evolve.
The study was published in Nature Communications.
Related topics
Antibiotics, Bacteriophages, Drug Discovery, Drug Discovery Processes, Peptide Therapeutics, Protein
Related organisations
Texas A&M University
Related people
Dr Karthik Chamakura, Dr Ryland Young