Whole animal screening: back to the future
Posted: 8 March 2017 | Horst Flotow ( Hit Discovery Constance (HDC) GmbH.) | No comments yet
High throughput screening (HTS) has become an integral and productive part of early-stage drug discovery, delivering hits that were the starting points for hit-to-lead programmes and resulting in a number of marketed drugs. HTS can be carried out in a number of formats, ranging from simple biochemical (target-based) to whole animal screening assay formats…


Generally, the simpler biochemical and whole cell assay formats have higher throughput, but lower biological relevance, as summarised in Figure 1. Thus, while the hits can be delivered faster with the simpler assays, they are also more likely to fail at the later stages of hit-to-lead or lead optimisation programmes.
This suggests that hits from biochemical, target-based screens in particular should be evaluated more rigorously in orthogonal and secondary assays before entering into hit-to-lead and lead optimisation. On the other hand, the slower but higher biological relevance screening with phenotypic whole cell, or even whole animal assays, can yield hits that pass muster in orthogonal and secondary assays, but may require more extensive target deconvolution and confirmation of the mechanism of action before hit-to-lead or lead optimisation chemistry can commence. Weighing up the relative benefits of throughput versus biological relevance is not always straightforward. However, it has also become clear that phenotypic assays provide enormous benefits in drug discovery, particularly in discovering first-in-class or novel drugs.1 While HTS in traditional animal models (e.g, rodents) is neither practicable, simple or inexpensive, animal models that provide a workable compromise between throughput and biological relevance are now available.
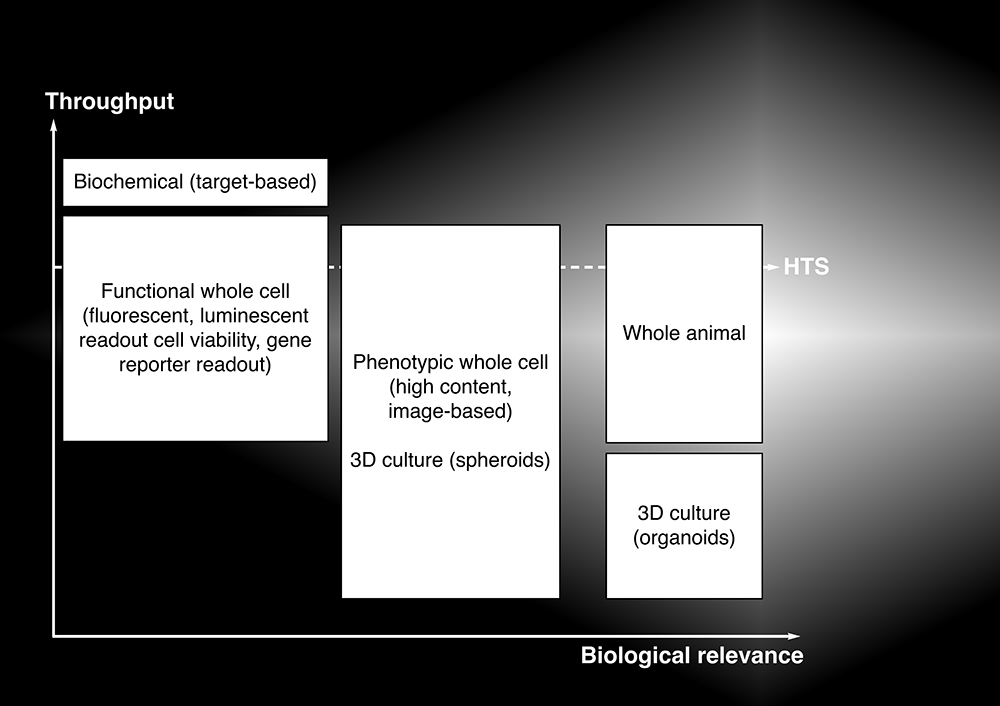

Figure 1: Relationship between throughput and biological relevance for different assay formats
Swim or wriggle?
There are two animal models that appear to be particularly suitable for HTS: the vertebrate zebrafish and the nematode Caenorhabditis elegans. Both are biologically well-characterised small-animal model systems, with extensive genetic information available. While adult zebrafish are too large to fit into microtitre plates, their embryos easily fit into 96 well plates. In addition, the embryos are transparent and compounds can be tested by simply adding them to the water in the wells. The phenotypic effects of the compounds on most of the major organs can then be visually determined.2 C. elegans is a small (up to 1mm long) and optically transparent nematode. Like zebrafish, it is a well-characterised model system, especially for developmental biology. C. elegans is cheap and easy to maintain in the laboratory and can be grown in 96, 384, or even 1536 well, microtitre plates for high throughput screening.3 Over the last few years HTS technologies have progressed in leaps and bounds, with image-based screening – also known as high content screening (HCS) – being at the leading edge. With the application of HCS analysis pipelines to zebrafish embryos recently, several examples have emerged that demonstrate the feasibility of doing in vivo screening (albeit in 96 well plates, with moderate throughput) with this vertebrate.4,5 However, the image acquisition and analysis remains a big challenge for a complex vertebrate like zebrafish embryos. To develop robust HTS and HCS in whole animal systems, investigators are increasingly turning in the direction of a simpler system, the nematode C. elegans.
C. elegans is very simple and convenient to culture on solid or liquid media, with bacteria such as E. coli as a food source, and the worms can be grown in sufficient quantities to be able to run HTS. In addition, a number of fluorescently labelled strains have been generated that can be used for screening.6 Some examples of anti-infective (antibiotics) and anti-cancer HTS using C. elegans are discussed below.
The worm has turned
The basic assay principle underpinning the use of C. elegans in anti-infective drug discovery is as follows. The normal food source for C. elegans is (non-pathogenic) bacteria such as E. coli. The worms ingest and digest the bacteria without being overwhelmed by them. However, when cultured in the presence of a number of pathogenic bacteria and fungi, for example, Enterococcus faecalis, Burkholderia pseudomallei, or Candida albicans, the nematodes are overwhelmed and die.7-10 This effect can be reversed by compounds such as antibiotics that kill the bacteria or fungi (or render them non-pathogenic), or by downregulation/deletion of genes involved in bacterial virulence or pathogenicity. Therefore C. elegans is a very sensitive and convenient whole animal model system that can be used in antibiotic and antifungal drug discovery, as outlined in some of the following examples.


There is an urgent need to find antibiotics that have a novel mechanism of action. The following example highlights the utility of a C. elegans bacterial infection model for identifying new and novel targets for HTS. The genomes of some of the human pathogens have been sequenced and libraries of mutant bacteria carrying deletions or mutations of many of the genes have been constructed.
Garvis and co-workers used such a library of 2,200 Pseudomonas aeruginosa mutants and screened them against C. elegans in 96 well plates. The aim was to find bacterial genes that would enhance worm survival (by attenuating the virulence of the bacteria). In their screen they identified several proteins in the P. aeruginosa chemotaxis pathway that appear to play a role in bacterial virulence or pathogenicity. These bacterial genes may therefore represent new targets for drug discovery.11 It may also be possible to use this approach to elucidate the mechanisms of action of novel antibiotic compounds (target deconvolution) found in whole animal or phenotypic HTS.
The next step in the drug discovery process would be HTS. Using E. faecalis-infected C. elegans, just over 37,000 compounds were screened in a 384 well microtitre plate assay. To measure nematode survival, Sytox Orange staining of dead worms and automated image analysis were used. The Sytox Orange only stains dead worms, thus allowing a quantification of the fraction of dead worm and identification of hit compounds that enhance survival of the worms. As the shape of the worms is also diagnostic for whether they are dead or alive, the screen found compounds that enhanced worm survival (presumably by acting in a bactericidal or bacteriostatic fashion).7 This is one of the first examples of the application of HCS using C. elegans, with a still modest throughput.
More recently, Lakshmanan et al used a fluorescent strain of C. elegans to develop an infection model of melioidosis. Melioidosis is an infectious disease endemic to Southeast Asia and Northern Australia, which is caused by a gram-negative bacterium, Burkholderia pseudomallei. Being a gram-negative organism, treatment options are limited and the severe cases of melioidosis have very high mortality rates (greater than 30%, depending on the area) so there is an urgent medical need for new treatments. Since the causative organism is a class B bioterrorism agent (and therefore needs to be handled in Biosafety Level 3, or BSL3 containment), the screening assay was developed using a closely related organism, Burkholderia thailandensis (which is less virulent, rarely causes human disease, and can be handled in a BSL2 facility).
The assay principle was simple. Sytox Green staining was used to quantify the number of dead worms and hits scored as wells containing few, if any, dead worms. The assay was run in 384 well plates and could be read in an imaging plate reader (PerkinElmer Viewlux) giving it very high throughput and allowing an HTS against a 300,000 compound library to be performed. It should be noted here that the control compound used on each plate during the HTS was co-trimoxazole, which is a clinical treatment for melioidosis, so this screening assay recapitulated at least some of the characteristics of the disease. The relevance of the C. elegans screening assay as a melioidosis model was further validated with some of the hits from the screen being active against clinical isolates of Burkholderia pseudomallei.8
Nematodes (helminths) themselves can also be important pathogens, particularly in agriculture but also in humans. As in the case of antibiotics (where the bacteria are developing resistance to current treatments), the nematodes are also developing resistance to currently used pesticides and treatments, and there is an urgent need to discover novel anthelmintic compounds. C. elegans is an ideal model system for this kind of screening, as shown in the anti-infective drug discovery examples above. Leung et al have constructed a dual fluorescence reporter assay to measure the activity of a gene regulatory pathway involved in the worms’ response to certain environmental compounds, as well as cell lineage determination. This C. elegans assay was adapted to run in 1536 well plates to increase the throughput (a format referred to as ultra HTS). The compounds found in the primary screen were subjected to several secondary screens in order to sift out compounds that were simply toxic or had a non-specific interaction with elements of the assay.12
The extensive genetic characterisation of C. elegans has shown that it contains many of the same signalling pathways found in humans, including many suspected of involvement in cancer. Recent developments in genetic engineering of the worms has resulted in the construction of numerous strains that read out the activity (or otherwise) of some of the more important pathways and targets, such as cell death and EGFR, with the basic assay principle being a gene reporter readout (fluorescence or luminescence) similar to a functional whole cell assay.
C. elegans is of particular use to developmental biologists, who have mapped the fate of most of the worm’s cells during its development. For example, the involvement of the EGFR signalling pathway in the development of the worm’s vulva has been well-characterised. Given the importance of EGFR tyrosine kinase inhibitors in treating a number of human cancers, the worms lend themselves as a model system to use for the screening of novel EGFR pathway inhibitors. Using C. elegans expressing human EGFR tyrosine kinase, a screen against a small compound library found known tyrosine kinase inhibitors, such as gefitinib and erlotinib, as well as new compounds that target downstream components of the pathway.13 Because the EGFR pathway and its role in the worms’ development has been so well characterised (with numerous mutants available), the target deconvolution can be relatively swiftly accomplished as well, and targets or mechanisms of action assigned to new compounds identified during the screening.
Conclusions and perspectives
Screening using whole animal models in infectious disease and cancer has made impressive progress in recent years, especially using the nematode worm C. elegans. Using complex whole animal model systems for HTS might seem counterintuitive. However, one has the ability to engineer a whole animal system and to target the assay to a specific pathway or target within the pathway. It is also clear that there is a great deal of additional information that can be obtained from such assays, especially information about potential compound toxicity. Although not as closely related to humans as zebrafish, the convenience and throughput of worm screening allows for a very fast and cost-effective screening of vast compound libraries.
In addition, the ease with which one can construct a highly target-specific assay in C. elegans means that it could be used, for example, to screen for inhibitors of drug resistant mutants of kinases and other enzyme targets, not just in a cellular but in a whole animal context. Conversely, if the original screen is less targeted (for example, a simple phenotypic screening assay), there are viable and timely alternative approaches and strategies to define the target in the bacteria (by screening compound activity against bacterial mutants) or in C. elegans (by screening against genetic mutants of the worms).
While C. elegans-based whole animal assays are ready to HTS now, zebrafish assays for HTS will need further development. This is an exciting area of assay development that will have a big impact on drug discovery in the next few years.
Biography


HORST FLOTOW is CEO at Hit Discovery Constance (HDC) GmbH.
References
- Zheng W, Thorne N, McKew JC. Phenotypic screens as a renewed approach for drug discovery. Drug Discovery Today. 2013 Nov 1;18(21-22): 1067–73
- Binder V, Zon LI. High throughput in vivo phenotyping: The zebrafish as tool for drug discovery for hematopoietic stem cells and cancer. Drug Discovery Today: Disease Models. 2013 Sep 2;10(1): e17–e22
- Leung MCK, Williams PL, Benedetto A, Au C, Helmcke KJ, Aschner M, et al. Caenorhabditis elegans: An emerging model in biomedical and environmental toxicology. Toxicological Sciences. 2008 Aug 18;106(1): 5–28
- Arulmozhivarman G, Stöter M, Bickle M, Kräter M, Wobus M, Ehninger G, et al. In vivo chemical screen in zebrafish embryos identifies regulators of hematopoiesis using a semiautomated imaging assay. Journal of Biomolecular Screening. 2016 Sep 20;21(9): 956–64
- Westhoff JH, Giselbrecht S, Schmidts M, Schindler S, Beales PL, Tönshoff B, et al. Development of an automated imaging pipeline for the analysis of the zebrafish larval kidney. Hwang S-PL, editor. PLoS ONE. 2013 Dec 4;8(12): e82137–13
- Leung CK, Deonarine A, Strange K, Choe KP. High-throughput screening and biosensing with fluorescent C. elegans strains. JoVE. 2011;(51)
- Moy TI, Conery AL, Larkins-Ford J, Wu G, Mazitschek R, Casadei G, et al. High-Throughput Screen for Novel Antimicrobials using a Whole Animal Infection Model. ACS Chem Biol. 2009 Jul 17;4(7):527–33
- Lakshmanan U, Yap A, Fulwood J, Yichun L, Hoon S, Lim J, et al. Establishment of a Novel Whole Animal HTS Technology Platform for Melioidosis Drug Discovery. Comb Chem High Throughput Screen. 2014 Nov 7;17(9):790–803
- Okoli I, Coleman JJ, Tempakakis E, An WF, Holson E, Wagner F, et al. Identification of Antifungal Compounds Active against Candida albicans Using an Improved High-Throughput Caenorhabditis elegans Assay. Idnurm A, editor. PLoS ONE. Public Library of Science; 2009 Sep 14;4(9):e7025–8
- Breger J, Fuchs BB, Aperis G, Moy TI, Ausubel FM, Mylonakis E. Antifungal Chemical Compounds Identified Using a C. elegans Pathogenicity Assay. PLoS Pathog. 2007;3(2):e18–11
- Garvis S, Munder A, Ball G, de Bentzmann S, Wiehlmann L, Ewbank JJ, et al. Caenorhabditis elegans Semi-Automated Liquid Screen Reveals a Specialized Role for the Chemotaxis Gene cheB2 in Pseudomonas aeruginosa Virulence. Ausubel FM, editor. PLoS Pathog. 2009 Aug 7;5(8): e1000540
- Leung CK, Wang Y, Malany S, Deonarine A, Nguyen K, Vasile S, et al. An Ultra High-Throughput, Whole-Animal Screen for Small Molecule Modulators of a Specific Genetic Pathway in Caenorhabditis elegans. Lamitina T, editor. PLoS ONE. 2013 Apr 29;8(4): e62166–12
- Bae Y-K, Sung JY, Kim Y-N, Kim S, Hong KM, Kim HT, et al. An In Vivo C. elegans Model System for Screening EGFR-Inhibiting Anti-Cancer Drugs. Hart AC, editor. PLoS ONE. 2012 Sep 5;7(9): e42441
Related topics
Assays, Drug Discovery, High-Throughput Screening (HTS), Hit-to-Lead, Screening