Translating stem cell biology into drug discovery
Posted: 22 June 2016 | Ilyas Singeç (National Center for Advancing Translational Sciences) | No comments yet
Pluripotent stem cell research has made extraordinary progress over the past decade. The robustness of nuclear reprogramming of somatic cells into induced pluripotent stem cells (iPSCs) has created entirely novel opportunities for drug discovery and personalised regenerative medicine…
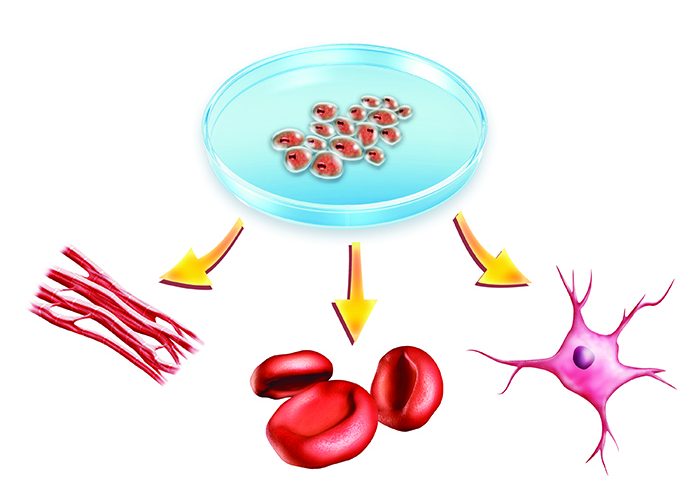

Patient- and disease-specific iPSCs can be expanded indefinitely and differentiated into relevant cell types of different organ systems. As the utilisation of iPSCs is becoming a key enabling technology across various scientific disciplines, there are still important challenges to be addressed. Here we review the current situation and reflect on the issues that the stem cell and translational communities are facing in bringing iPSCs closer to clinical application.
Quality assurance of iPSCs
Pluripotent stem cells are characterised by extensive self-renewal under appropriate cell culture conditions and differentiation into all cell types of the human body. The groundbreaking discovery by Shinya Yamanaka and colleagues1,2 over a decade ago that somatic cells can be reprogrammed into embryonic-like cells by defined factors has reinvigorated biomedical research in general and human biology-oriented drug discovery in particular. After quickly establishing that somatic cells – independent of tissue of origin, age and reprogramming technique – can be robustly reverted back and stably maintained at a pluripotent state3,4, the field has moved on to generating new iPSC lines from patients with monogenic and complex genetic diseases, as well as those of unknown etiology.
Studying relevant cell types in a dish in order to better understand the molecular underpinnings of intractable human diseases holds great promise with developing novel mechanism-based concepts in drug screening. To enable such disease modelling, predictive toxicology, and regenerative cell therapy applications, major international efforts and public-private partnerships are underway to generate large iPSC banks considering ethnic and disease-specific backgrounds5. While many new iPSC lines are being generated, important questions need to be answered in order to ensure acceptable standardisation, comprehensive characterisation, rigorous quality control and safety. Issues that need to be overcome include: the lack of experimental reproducibility, uncontrolled differentiation protocols, genome instability of reprogrammed and differentiated cells, and transplantation of unwanted cells with potential teratoma formation risk.
Recent work has demonstrated the equivalence of genetically matched iPSCs and embryonic stem cell lines (ESCs)6. This is an important comparison, since ESCs are still considered the gold standard for bona fide pluripotency. In addition, this study highlighted that genetic background can be a major confounding factor for comparative studies that rely solely on genetic and epigenetic signatures, as well as underscored the importance of being careful when drawing conclusions based on comparing only a few iPSC lines – currently the main approach in the field. Moreover, further systematic studies are necessary to establish criteria that can reliably distinguish ‘high-quality’ iPSCs from ‘poor-quality’ iPSCs.

Many new iPSC lines are being created, but issues such as quality control need to be addressed.
At present the most widely used parameters to characterise iPSCs include morphology, karyotype analysis, immunophenotyping (for example, surface makers, transcription factors), multilineage differentiation (that is, ectoderm, mesoderm, endoderm) by generating embryoid bodies and in vivo teratoma formation. Other approaches suggested as a measure of pluripotency include bioinformatics analysis based on gene expression7,8. However, whether these approaches are acceptable enough to completely replace teratoma formation is still being debated. Working with iPSCs is labour-intensive and costly, and identifying ‘high-quality’ iPSCs is a critical goal which should be prioritised as early as possible in the drug discovery process. Nevertheless, it remains unclear if the currently available assays and protocols are sufficient and rigorous enough for clinical and translational purposes.
Reproducible differentiation protocols
The remarkable biological versatility of pluripotent stem cells is based on their developmental potential to give rise to all somatic cell types, while not undergoing cellular senescence in the pluripotent state. Attempts to harness this potential of unlimited cell growth and differentiation began after the successful derivation of the first human ESC lines9. Several approaches have been used over the years to differentiate pluripotent cells into desired lineages and cell types, including overgrowing cells and culturing them as neurospheres10; forming free-floating embryoid bodies and picking neural rosettes11; co-culture with stromal cells12; using recombinant proteins (e.g., Noggin)13, small molecule inhibitors such as SB431542 alone14 or in combination with recombinant Noggin15.
While over the years there has been a trend to move from empirical methods to directed differentiation strategies by targeting specific cell signalling pathways, there are still major knowledge gaps and biological unknowns that hinder the formulation of highly reproducible differentiation protocols to yield large numbers of mature cell types at high purity (Figure 1). Questions remain. How can the stem cell field overcome the challenge of heterogeneous cultures with immature cell types generated over extended periods of time (weeks to months) and exposed to a flurry of small molecules and recombinant proteins, often used at supraphysiological concentrations? How can we comprehensively characterise cell type identities across different developmental stages? How can we identify and manipulate relevant cell signalling pathways and new small molecules that target these pathways? Bringing solutions to these questions is central to firmly establishing the iPSC technology in drug discovery and clinical applications.
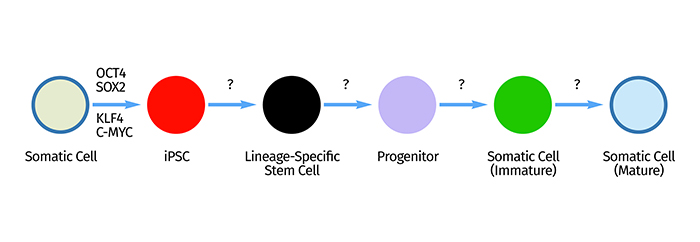

Figure 1: Reprogramming of somatic cells into iPSCs by transient expression of defined transcription factors (OCT4, SOX2, KLF4, C-MYC) is a robust and highly reproducible procedure. However, efficient and controlled multistep differentiation of iPSCs into transient phenotypes and mature functional cells and precise characterisation of these cell type identities across developmental states are currently among the greatest challenges in stem cell biology.
To tackle these widely accepted challenges, the National Institutes of Health (NIH), as part of its Regenerative Medicine Program, has recently launched the Stem Cell Translation Laboratory (SCTL). The SCTL is located within the National Center for Advancing Translational Sciences (NCATS) and is dedicated to bringing the iPSC technology closer to drug discovery and regenerative medicine applications (see https://commonfund.nih.gov/stemcells/index).
Automation and industrialised scale-up of iPSC cultures
Precisely formulating robust differentiation protocols requires deeper and actionable insights into the basic biology of pluripotent cells. The resulting information and reference datasets should then allow for better control of cell fate choice and terminal differentiation. To this end, it is critical to perform dose-response experiments, elucidate signal strengths, and carefully document the net effect of combinatorial manipulations of relevant pathways. The importance of combining genomic and proteomic methods is underscored by the fact that mRNA abundance often does not predict protein expression levels, particularly during dynamic transitions16. Integration and analysis of such complex orthogonal datasets by systems biology methods should enable establishing a roadmap that is practical and cost-efficient enough to be used across many different laboratories and translational research settings.
Routine manual cell culture work is inherently influenced by the physico-chemical environment, as well as human bias. Typically, scientists working in cell culture have their own style and preference in performing daily tasks. Variations in carrying out specific tasks, such as adding fresh medium at varying time points, cell passaging at varying cell densities, and other more or less significant or subtle effects can all have a strong impact on the experimental outcome and the quality of the end product. Standardising routine cell culture and the manufacturing process in cell engineering are critical steps, with automated robotic cell culture systems providing unique opportunities to establish standard operating procedures (SOPs), reduce human bias, and control cell culture conditions more precisely.
Using the robotic CompacT SelecT system17 combined with a chemically defined medium18, we have started to establish protocols that can maintain and expand pluripotent stem cells under highly controlled feeder-free conditions (Figure 2).
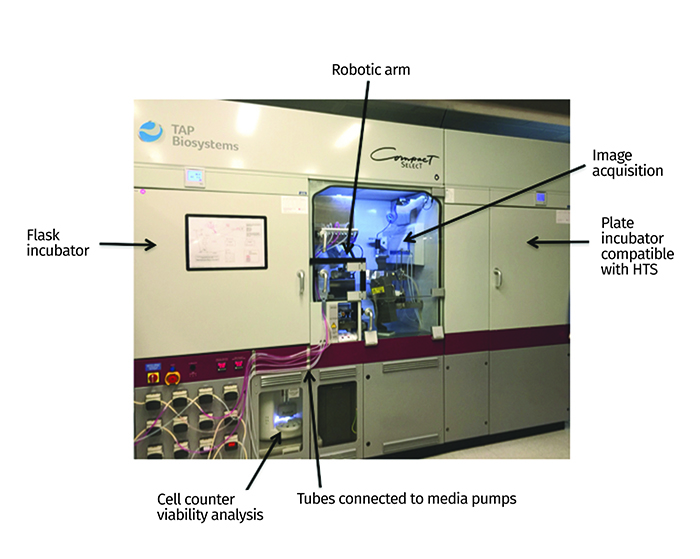

Figure 2: The CompacT SelecT automated cell culture platform allows standardisation and scale-up of iPSCs and their differentiated progeny. Some key features and components of the system are highlighted with arrows. HTS: high-throughput screening.
The fully automated workflow allows each step to be controlled via computer-aided commands. This real-time system ensures that the scientist can conveniently control and monitor cell culture around-the-clock. For instance, daily media change can be performed consistently at exact times. Any possible technical error is immediately transmitted to the scientist in charge. Integrated cell counter, viability assessment, and image-based passaging are key features for standardisation and documentation. In summary, automation provides the following advantages that will help to implement standardised utilisation of iPSCs for translational research:
- Expansion and maintenance of multiple cell lines
- Cell counting and viability measurement
- Sub-culturing and cell passaging
- Cell seeding
- Transient transfection
- Harvesting at exact time points
- Reducing the risk of cell culture contamination
- Incubation of up to 182 T-flasks and up to 420 plates in current configuration
- Plating into 96, 384 and 1536-well plates suitable for high-throughput and high-content screening.
Industrialised scale-up and access to large numbers of properly differentiated cell types will impact future health care by leveraging both personalised medicine and treatment of large patient cohorts.


Scientists working in cell culture carry out specific tasks in varying ways.
Transforming drug discovery by human biology
The traditional drug discovery process is painstakingly slow and inefficient19-21. The high attrition rate and failure of potential drugs due to safety issues and lack of efficacy in patients detected only after many years of investment suggest it is critical to re-visit the usefulness of preclinical models that have been relied upon in the past. Indeed, overdependence on animal models and immortalised cell lines in the preclinical stage is very risky and often times of limited predictive value for the outcome in the clinical phase22-25. More than 90% of drugs developed based on animal models fail in clinical trials23. To address at least some of the obstacles, drug repurposing has emerged as a valuable strategy. The underlying concept, which is to find new indications for existing drugs, can avoid redundant and costly preclinical studies and Phase 1 safety trials21. Along these lines, the NCATS has compiled the largest public repository of approved and clinical phase drugs – the NCATS Pharmaceutical Collection (NPC) – aimed at broadly sharing this resource for drug repositioning, toxicology studies and chemical genomic profiling26, 27.
The two main strategies of small molecule drug discovery can be classified as either target-based or phenotypic28. While target-based approaches aim to modulate a specific molecular target of interest, the phenotypic approach is intended to modify a disease-associated phenotype in cells or whole organisms. Comparing the success rate of both approaches has led to a renaissance of phenotypic screens28. However, setting up informative phenotypic screens requires a renewable cell source and validation of cellular assays that faithfully recapitulate human physiology. Patient- and disease-specific iPSCs are uniquely suited for developing novel human biology-focused paradigms for drug discovery and first-in-class therapies.
Once the challenges discussed herein are sufficiently addressed (i.e., quality assurance, reproducible and scalable differentiation), iPSC-based models will be broadly implemented in all stages of preclinical drug development. For instance, high-throughput and high-content screening, hit validation, structure-activity relationship studies, and toxicology assays will directly benefit from the consistency of utilising the same cell types(s) derived from genotyped patients with a known clinical history. A few studies have started to employ pluripotent cells for drug screening29-32. As we learn to robustly differentiate and scale up iPSC-derived cells, it is apparent that more systematic and extensive screening studies using much larger compound libraries will follow in the near future.
Beyond using human cells in screening, careful analysis of iPSC-based cellular assays will also provide insights into the complexity of human disease, leverage the understanding of genotype-phenotype relations and pinpoint novel cellular and molecular disease mechanisms33-36. Elucidating genetic diseases, particularly those with underlying strong cell-autonomous effects, may indeed enable robust and streamlined identification and validation of new ‘druggable’ targets, signalling pathways and disease phenotypes.
In a different approach, the wealth of genetic data obtained by genome-wide association studies (GWAS) can be interrogated by probing disease-relevant cell types. Follow-up experiments and prioritisation of hundreds of potential disease targets based on GWAS data is a significant problem. This is further complicated by the fact that the majority of hits are found in regulatory elements of unknown significance and not in protein-coding regions37. It is, again, desirable that these targets be directly investigated in relevant human cells from affected patients under defined laboratory conditions. The versatility of the iPSC technology (Figure 3) can be further enhanced by the power of functional genomics (e.g., RNAi) and genome editing such as the CRISPR/Cas9 system, which allows precise site-specific genetic manipulations. The interrogation of iPSC-based micro-physiological systems (‘tissue-chips’) and three-dimensional model systems38 will bring exciting novel opportunities, too. Together, these cell and gene engineering technologies will transform modern day drug discovery and also shed new light on designing successful clinical trials.
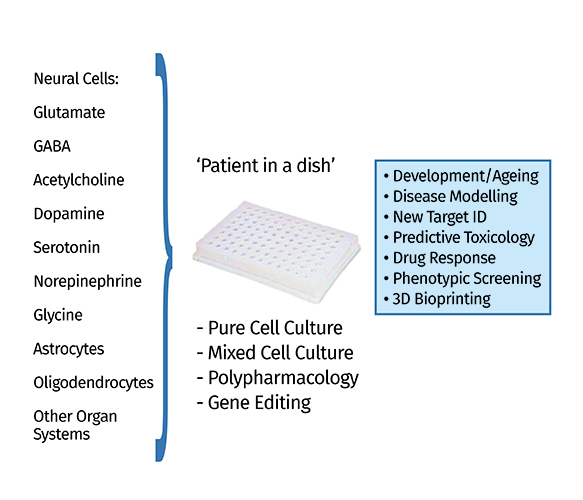

Figure 3: Summary of the many biomedical opportunities enabled by the iPSC technology (‘patient in a dish’). For instance, after generating specific neuronal subtypes representative of various neurotransmitter systems (e.g., glutamatergic, GABAergic, dopaminergic), these cells can be investigated as pure and mixed cultures and subjected to various manipulations (e.g., polypharmacology) for new target identification, predictive toxicology and other purposes.
Conclusion
The discovery of iPSCs and the work of the past decade have moved this remarkable technology closer to drug screening and clinical applications. Improving the robustness of differentiation protocols and implementing SOPs for scale-up and standardised assays will firmly establish the iPSC technology across many different translational communities. Such an integrated precision medicine strategy that is genetics-based, cell type-specific and clinical data-informed will enable human diseases to be interrogated and treated following the principles of evidence-based medicine. Innovative new therapeutics based on truly novel disease mechanisms should eventually create a return on investment and significantly increase the efficiency of drug discovery for common and rare diseases.
Acknowledgement
We are grateful to Pınar Ormanoğlu and Steven Titus for their excellent ongoing support. We thank all our colleagues at NCATS and the NIH Common Fund (Regenerative Medicine Program) for their collaboration and commitment to help patients.
Biographies




References
- Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006; 126(4):663-76
- Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007; 131(5):861-72
- Aoi T, Yae K, Nakagawa M, Ichisaka T, Okita K, Takahashi K, Chiba T, Yamanaka S. Generation of pluripotent stem cell from adult mouse liver and stomach cells. Science. 2008; 321(5889):699-702
- Okita K, Nakagawa M, Hyenjong H, Ichisaka T, Yamanaka S. Generation of mouse induced pluripotent stem cells without viral vectors. Science. 2008; 322(5903):949-53
- Wilmut I, Leslie S, Martin NG, Peschanski M, Rao M et al. Development of a global network of induced pluripotent stem cell haplobanks. Regen. Med. 2015; 10(3):235-8
- Choi J, Lee S, Mallard W, Clement K, Tagliazucchi GM, Lim H, Choi IY, Ferrari F, Tsankov AM, Pop R, Lee G, Rinn JL, Meissner A, Park PJ, Hochedlinger. A comparision of genetically matched cell lines reveals the equivalence of human iPSCs and ESCs. Nat. Biotechnol. 2015; 33(11):1173-81
- Müller FJ, Laurent LC, Kostka D, Ulitsky I, Williams R, Lu C, Park IH, Rao MS, Shamir R, Schwartz PH, Schmidt NO, Loring JF. Regulatory networks define phenotypic classes for human stem cell lines. Nature. 2008; 455(7211):401-5
- Tsankov AM, Akopian V, Pop R, Chetty S, Gifford CA, Daheron L, Tsankova NM, Meissner A. A qPCR ScoreCard quantifies the differentiation potential of human pluripotent stem cells. Nat. Biotechnol. 2015; 33(11):1182-92
- Thomson JA, Itskovitz-Eldor J, Shapiro SS, Waknitz MA, Swiergiel JJ, Marshall VS, Jones JM. Science. 1998; 282(5391):1145-7
- Reubinoff BE, Itsykson P, Turestsky T, Pera MF, Reinhartz E, Itzik A, Ben-Hur T. Neural progenitors from human embryonic stem cells. Nat. Biotechnol. 2001; 19(2):1134-40
- Zhang SC, Wernig M, Duncan ID, Brüstle O, Thomson JA. In vitro differentiation of transplantable neural precursors from human embryonic stem cells. Nat. Biotechnol. 2001; 19(12):1129-33
- Perrier AL, Tabar V, Barberi T, Rubio ME, Bruses J, Topf N, Harrison NL, Studer L. Derivation of midbrain dopamine neurons from human embryonic stem cells. Proc. Natl. Acad. Sci. USA 2004; 101(34):12543-8
- Pera MF, Andrade J, Houssami S, Reubinoff B, Trounson A, Stanley EG, Ward-van Oostwaard D, Mummery C. Regulation of human embryonic stem cell differentiation by BMP-2 and its antagonist noggin. J. Cell Sci. 2004; 117:1269-80
- Smith JR, Vallier L, Lupo G, Alexander M, Harris WA, Pedersen RA. Inhibition of Activin/Nodal signaling promotes specification of human embryonic stem cells into neuroectoderm. Dev. Biol. 2008; 313(1):107-17
- Chambers SM, Fasano CA, Papapetrou EP, Tomishima M, Sadelain M, Studer L. Highly efficient neural conversion of human ES and iPS cells by dual inhibition of SMAD signaling. Nat. Biotechnol. 2009; 27(3):275-80
- Liu Y, Beyer A, Aebersold R. On the dependency of cellular protein levels on mRNA abundance. Cell. 2016; 165(3):534-50
- Thomas RJ, Anderson D, Chandra A, Smith NM, Young LE, Williams D, Denning C. Automated, scalable culture of human embryonic stem cells in feeder-free conditions. Biotechnol. Bioeng. 102(6):1636-44
- Chen G, Gulbranson DR, Hou Z, Bolin JM, Ruotti V, Probasco MD, Smuga-Otto K, Howden SE, Diol NR, Propson NE, Wagner R, Lee GO, Antosiewicz-Bourget J, Teng JM, Thomson JA. Chemically defined conditions for human iPSC derivation and culture. Nat. Methods 2011; 8(5):424-9
- Pangalos MN, Schechter LE, Hurko O. Drug development for CNS disorders: strategies for balancing risk and reducing attrition. Nat. Rev. Drug Discov. 2007; 6(7):521-32
- Scannell JW, Blanckley A, Boldon H, Warrington B. Diagnosing the decline in pharmaceutical R&D efficiency. Nat. Rev. Drug Discov. 2012;11(3):191-200
- Tobinick EL. The value of drug repositioning in the current pharmaceutical market. Drug News Perspect. 2009; 22(2):119-25
- Nestler EJ, Hyman SE. Animal models of neuropsychiatric disorders. Nat. Neurosci. 2010; 13(10):1161-9
- van der Worp HB, Howells DW, Sena ES, Porritt MJ, Rewell S, O’Collins V, Macleod MR. Can animal models of disease reliably inform human studies? PLoS Med 2010; 7(3):e1000245
- Seok J, Warren HS, Cuenca AG, Mindrinos MN, Baker HV, et al. Genomic responses in mouse models poorly mimic human inflammatory diseases. Proc. Natl. Acad. Sci. USA 2013; 110(9):3507-12
- O’Connor MD. The 3R principle: advancing clinical application of human pluripotent stem cells. Stem Cell Res. Ther. 2013; 4(2):21
- Huang R, Southall N, Wang Y, Yasgar A, Shinn P, Jadhav A, Nguyen DT, Austin CP. The NCGC pharmaceutical collection: a comprehensive resource of clinically approved drugs enabling repurposing and chemical genomics. Sci. Transl. Med. 2011;3(80):80ps16
- Lee JA, Shinn P, Jaken S, Oliver S, Willard FS, et al. Novel Phenotypic outcomes identified for a public collection of approved drugs from a publicly accessible panel of assays. PLoS One 2015; 10(7):e)130796
- Swinney DC, Anthony J. How were new medicines discovered? Nat. Rev. Drug Discov. 2011; 10(7):507-19
- Makhortova NR, Hayhurst M, Cerqueira A, Sinor-Anderson AD, Zhao WN et al. A screen for regulators of survival of motor neuron protein levels. Nat. Chem. Biol. 2011; 7(8):544-52
- Lee G, Ramirez CN, Kim H, Zeltner N, Liu B et al. Large-scale screening using familial dysautonomia induced pluripotent stem cells identifies compounds that rescue IKBKAP expression. Nat. Biotechnol. 2012; 30(12):1244-8
- Choi SM, Kim Y, Shim JS, Park JT, Wang RH et al. Efficient drug screening and gene correction for treating liver disease using patient-specific stem cells. Hepatology 2013; 57(6):2458-68
- Charbord J, Poydenot P, Bonnefond C, Feyeux M, Casagrande F et al. High throughput screening for inhibitors of REST in neural derivatives of human embryonic stem cells reveals a chemical compound that promotes expression of neuronal genes. Stem Cells 2013; 31(9):1816-28
- Grskovic M, Javaherian A, Strulovici B, Daley GQ. Induced pluripotent stem cells—opportunties for disease modeling and drug discovery. Nat. Rev. Drug Discov. 2011; 10(12):915-29
- Matsa E, Burridge PW, Wu JC. Human stem cells for modeling heart disease and for drug discovery. Sci. Transl. Med. 2014; 6(239):239ps6
- Kimbrel EA, Lanza R. Current status of pluripotent stem cells: moving the first therapies to the clinic. Nat. Rev. Drug Discov. 2015;14(10):681-92
- Plenge RM, Scolnick EM, Altshuler D. Validating therapeutic targets through human genetics. Nat. Rev. Drug Discov. 2013; 12(8):581-94
- Schizophrenia Working Group of the Psychiatric Genomics Consortium. Biological insights from 108 schizophrenia-associated genetic loci. Nature 2014; 511(7510):421-7
- Schwartz MP, Hou Z, Propson NE, Zhang J, Engstrom CJ et al. Human pluripotent stem cell-derived neural constructs for predicting neural toxicity. Proc. Natl. Acad. Sci. USA 2015; 112(40):12516-21
Related topics
Drug Discovery, Screening, Stem Cells, Translational Science