Newly identified compound reduces potency of Shiga toxin
Posted: 5 August 2022 | Ria Kakkad (Drug Target Review) | No comments yet
Scientists have identified a molecule that can bind to a lethal subtype of the bacterial Shiga toxin, suppressing its deadly effects.

A strain of E. coli bacteria called enterohemorrhagic E. coli (EHEC) is known to cause several gastrointestinal disorders, which include bloody diarrhoea and abdominal cramps, by damaging the intestinal lining. When accompanied with fatal systemic complications, it can even cause acute renal failure in children. The EHEC exerts these deadly effects by producing the Shiga toxin (Stx), of which the Stx2a subtype is particularly virulent and deadly. Compounds that can inhibit these toxins are, therefore, desirable as potential therapeutics against EHEC infections.
Scientists from Doshisha University, Japan has recently discovered a molecule that inhibits Stx2a toxicity by binding to its “A-subunit” – the part of the toxin responsible for its lethality. The study was recently published in Scientific Reports.
“The catalytic A-subunit of Stx2a toxin inhibits protein synthesis and its inhibition could be crucial for slowing EHEC pathogenesis,” explained Professor Kiyotaka Nishikawa, a scientist who led the study.
In their research, the team identified the potent compound from a database with over 7,400,000 molecules. To do this, they first had to identify the basic three-dimensional (3D) arrangement of molecules that can occupy the catalytic cavity in the A-subunit. The researchers were able to find synthetic molecule with a high affinity for A-subunit, a peptide called “MMβA-mono,” helped identify the compound that could bind to the A-subunit of Shiga toxin by serving as a template.
The researchers next outlined the molecular and electronic structure that a possible inhibitory compound must have using structural analysis and X-ray crystallography. These features of a potential inhibitor, known as a “pharmacophore,” was then confirmed using molecular dynamics simulations.
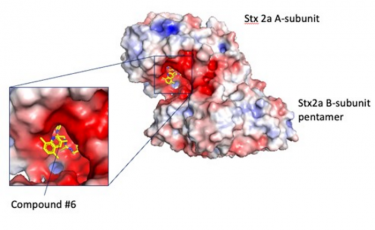

Researchers from Japan used a peptide-based pharmacophore to identify a potent molecule that binds to and inhibits the bacterial Shiga toxin, Stx2a. Results suggest that the novel compound #6 may be an effective therapeutic agent against enterohemorrhagic E. coli (EHEC) infections
[Credit: Professor Kiyotaka Nishikawa from Doshisha University, Japan].
Finally, they screened a chemical database for compounds that resembled the pharmacophore and identified nine candidates using docking simulations. Of these, a compound identified as “compound #6” showed effective binding to the A-subunit of Stx2a.
Further in vitro cytotoxicity assays using Vero cells showed that compound #6 significantly reduced the destruction of cells caused by Stx2a. Additionally, mice models treated with a lethal dose of Stx2a and compound #6 survived longer than those injected with only Stx2a.
“The hydrophobicity of compound #6 may facilitate penetration through the cell envelope, allowing it to inhibit the toxin present in the cells. We believe that it holds promise as a novel therapeutic agent for treating EHEC infections,” concluded Nishikawa.
Related topics
Assays, In Vitro, Molecular Biology, Molecular Targets, Screening
Related conditions
gastrointestinal disorders
Related organisations
Doshisha University
Related people
Professor Kiyotaka Nishikawa