Appearance of proteins used to predict function with machine learning
Posted: 10 December 2019 | Victoria Rees (Drug Target Review) | No comments yet
Researchers have used a machine-learning algorithm to study protein appearance and discover common features that influence function, which could be used to design artificial cells.
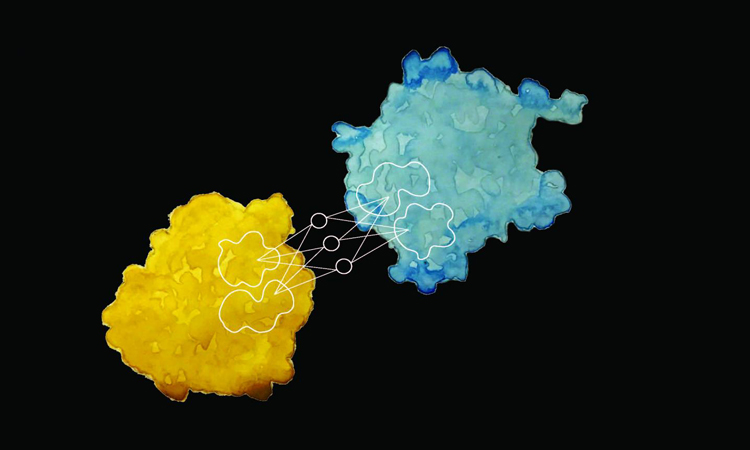

Researchers at EPFL have developed a new way to predict a protein's interactions with other proteins and biomolecules and its biochemical activity, merely by observing its surface (credit: Laura Persat / 2019 EPFL).
A new machine learning-driven technique has been able to predict the interactions between proteins and describe biochemical activity based on surface appearance.
The study was conducted at the Laboratory of Protein Design & Immunoengineering (LPDI), Switzerland, in collaboration with other researchers.
According to the team, the method, known as MaSIF, could support the development of protein-based components for artificial cells in novel therapeutics.
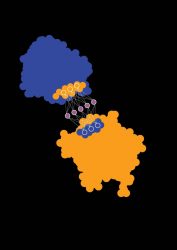

Scientists have developed a new way to predict a protein’s interactions with other proteins and biomolecules and its biochemical activity, merely by observing its surface (credit: Laura Persat / 2019 EPFL).
The researchers took a vast set of protein surface data and fed the chemical and geometric properties into a machine-learning algorithm, training it to match these properties with particular behaviour patterns and activity. They used the remaining data to test the algorithm.
“By scanning the surface of a protein, our method can define a fingerprint, which can then be compared across proteins,” says Pablo Gainza, the first author of the study.
The team found that proteins performing similar interactions share common features.
“The algorithm can analyse billions of protein surfaces per second,” says LPDI director Bruno Correia. “Our research has significant implications for artificial protein design, allowing us to program a protein to behave a certain way merely by altering its surface chemical and geometric properties.”
The method could also be used to analyse the surface structure of other types of molecules, say the researchers.
The findings were published in Nature Methods.
Related topics
Artificial Intelligence, Imaging, Protein, Research & Development
Related organisations
Laboratory of Protein Design & Immunoengineering (LPDI)
Related people
Bruno Correia, Pablo Gainza