Novel method of labelling DNA bases developed for sequencing
Posted: 19 May 2021 | Victoria Rees (Drug Target Review) | No comments yet
Using electrochemical detection and redox labels, researchers have created a new, faster method to sequence DNA.


A research team has developed a novel method for labelling DNA, which in the future could be used for sequencing DNA by means of electrochemical detection. The technique was created at the Czech Academy of Sciences (IOCB Prague), Charles University, both Czech Republic, and Universitat Rovira i Virgili (URV), Spain.
The team explain that the genetic information carried within a DNA molecule is determined by the order of its nucleotides. Despite great progress in recent years, current DNA sequencing methods, typically based on fluorescent labelling, are still time-consuming and relatively expensive techniques, so have some limitations. Therefore, scientists are intensively searching for new approaches to simplify and accelerate sequencing.
One promising approach is the use of electrochemical detection and so-called redox labels, which are compounds that can be oxidised or reduced on electrodes.
In their study, the team succeeded in designing and synthesising artificial nucleotides with special attached redox labels that can be oxidised on a gold or carbon electrode at a specific potential to produce a measurable and analytically useful signal. These labels are carboranes, cage structures composed of boron and carbon atoms, into which other metal atoms can be incorporated, such as iron or cobalt, affecting their resulting electrochemical properties.
The modified nucleotides have been engineered so that the enzyme DNA polymerase, which uses available nucleotide building blocks to build DNA within a cell, can incorporate them into a newly synthesised DNA strand. The researchers prepared a strand of DNA comprising these modified nucleotides, with each of the four types of nucleotide carrying its own unique label allowing for its subsequent identification.
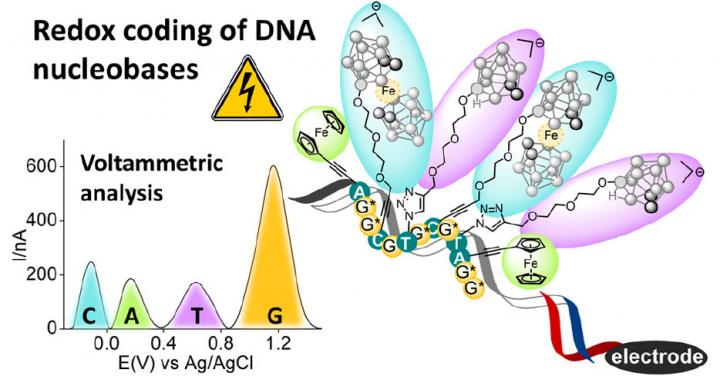

Redox coding of DNA nucleobases [credit: IOCB Prague].
Until now, researchers have only managed to label and measure one, at most two, types of redox-labelled nucleotides in a single strand of DNA. As each of the new modified nucleotides carries its own label, which during electrochemical detection gives a specific oxidation signal at varying potentials, the individual types of nucleotides can be distinguished. Moreover, the size of each signal is dependent on the number of copies of the given nucleotide in the DNA, which then makes it possible to quickly determine the relative representation of individual nucleotides in the measured DNA.
The researchers say the newly developed electrochemical coding of DNA bases offers a range of advantages, such as simpler and more affordable instrumentation and faster analysis. The team say the method holds promise for DNA sequencing and diagnostic applications.
The research is published in the Journal of the American Chemical Society.
Related topics
Analytical Techniques, DNA, Molecular Targets, Next-Generation Sequencing (NGS), Sequencing
Related organisations
Charles University, Czech Academy of Sciences (IOCB Prague), Universitat Rovira i Virgili (URV)