New approach improves avoidance of ‘bystander’ edits in CRISPR-base editor treatments
Posted: 15 November 2021 | Victoria Rees (Drug Target Review) | No comments yet
Rice University scientists have refined specific CRISPR-base editing strategies to avoid errors that occur during gene editing.

A new strategy by Rice University, US, scientists has sought to avoid gene-editing errors by fine-tuning specific CRISPR-base editing strategies in advance. The researchers aim to build better base editors, molecular machines that target and fix faulty DNA at single-base resolution.
The team say that the molecular processes that base editors use to manipulate strands of DNA is cutting them where necessary and making way for replacement code. When it works, as it increasingly does to treat genetic diseases like sickle cell anaemia and some cancers, the editor only edits the intended nucleotide. However, when it does not work, it is because bystander edits can cause undesired effects.
The new strategy primarily seeks to eliminate wayward edits to bystanders, nucleotides adjacent to the base editor’s target. The lab of Xue Sherry Gao previously introduced tools to improve the accuracy of CRISPR-based edits of cytosine mutations up to 6,000-fold. For the new project, they engaged the lab of Anatoly Kolomeisky to help create a theoretical framework to eliminate trial and error in the design of a library of editors. These would better target mutations that cause disease while avoiding bystanders. In the process, the framework could help scientists better understand the chemical and physical processes that take place during base editing.
“Sherry and other experimental scientists already had results that worked,” said Kolomeisky, referring to an earlier paper, in which the lab used its editor to convert cytosines to thymines, correcting the DNA mutations while avoiding otherwise vulnerable cytosines upstream. “But despite these amazing developments, there has been no microscopic understanding of what we have to do with these protein systems to improve editing… We applied the model for that [previous] result and got some important parameters we then used to design what mutations and where are needed to get precise editing. Ultimately, this symbiosis of theory and experiment allows us to work in a smart way.”
The new strategy, outlined in Nature Communications, combines molecular dynamics simulations and stochastic (aka random) models that pinpoint the binding energies between molecules required to achieve maximum editing selectivity. Experiments in Gao’s lab validated the CRISPR results.
Critically, the framework includes a way to characterise the binding affinity between deaminases – enzymes that catalyse the removal of an amino group from a molecule – and single-stranded DNA (ssDNA). Ideally, they said, the deaminase stays on the ssDNA just long enough to complete the primary edit and releases before inadvertently editing a bystander site.
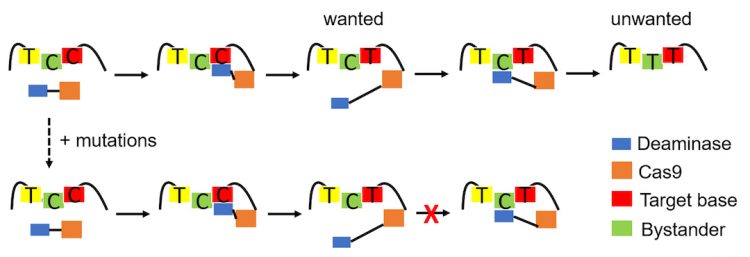
A new strategy by Rice University scientists seeks to avoid gene-editing errors by fine-tuning specific CRISPR-base editing strategies in advance. Their theoretical framework is intended to eliminate trial and error in the design of a library of editors [credit: Qian Wang/Kolomeisky Lab].
“The important thing here is that one mutation does not work for different systems,” Kolomeisky said. “So, for every system, you have to do this procedure again, but at least it is clear what should be done.”
“The model has been very successful in reflecting what has already been done experimentally,” Gao said. “But since then, we have been able to turn down bystander effects in other base-editing systems. Because the number of mutants could be in the thousands, it is unrealistic for experimentalists alone to verify individual base editors,” she said. “Only this multidisciplinary approach will allow us to build a huge library of editors computationally, then narrow the numbers down to the most promising candidates for further experimental verifications. That is what we are working toward.”
Related topics
CRISPR, DNA, Genome Editing, Precision Medicine
Related organisations
Rice University
Related people
Anatoly Kolomeisky, Xue Sherry Gao