ComFB protein discovery could help fight antibiotic resistance
Posted: 29 September 2025 | Drug Target Review | No comments yet
Scientists have discovered a new family of bacterial proteins – called ComFB – that regulate both movement and DNA uptake – suggesting potential new methods to combat pathogenic infections.

Bacteria use specialised motility structures, such as flagella or pili, not only for locomotion and colonisation but also to exchange DNA. This ability, known as genomic plasticity, plays a crucial role in bacterial pathogenicity, biofilm formation and the spread of antibiotic resistance.
A joint international effort
Through a collaborative effort involving researchers from, Heinrich Heine University Düsseldorf (HHU), the Universities of Tübingen, Gießen and Freiburg, the Max Planck Institute for Biology Tübingen, and colleagues from the US National Institutes of Health and New Jersey Medical School, scientists have identified a previously unknown family of signalling proteins. These proteins are abundant across bacteria and help regulate motility and DNA uptake mechanisms.
The findings were published in Cell Discovery and the Proceedings of the National Academy of Sciences (PNAS).
Biomarkers aren’t just supporting drug discovery – they’re driving it
FREE market report
From smarter trials to faster insights, this report unpacks the science, strategy and real-world impact behind the next generation of precision therapies.
What you’ll unlock:
- How biomarkers are guiding dose selection and early efficacy decisions in complex trials
- Why multi-omics, liquid biopsy and digital tools are redefining the discovery process
- What makes lab data regulatory-ready and why alignment matters from day one
Explore how biomarkers are shaping early drug development
Access the full report – it’s free!
ComFB protein: a key regulator
Led by Professor Dr Khaled Selim of the Institute of Phototrophic Microbiology at HHU, the research team discovered that second messenger molecules – c-di-GMP and c-di-AMP – interact with a newly identified receptor protein – ComFB – to regulate both bacterial mobility and DNA uptake.
“The ability of different ComFB proteins from various bacteria to bind and precisely integrate the motility/DNA uptake signal(s) is reported by the second messenger di-nucleotides (c-di-GMP and c-di-AMP),” said PhD student Sherihan Samir, first author of both publications.
These proteins were studied in cyanobacteria, Bacillus subtilis and Vibrio cholerae – the pathogen responsible for cholera outbreaks.
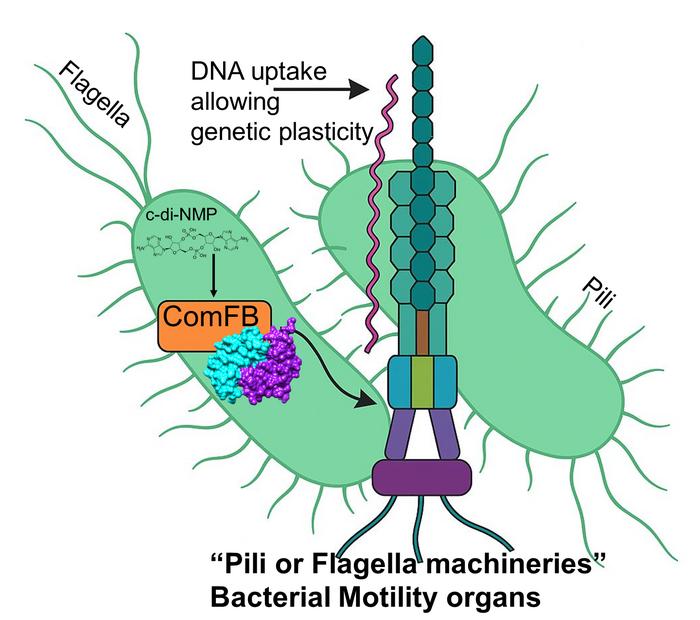

The contractile pilus mechanism, which enables the bacteria to move, is regulated by c-di-AMP signals via the receptor protein ComFB. This mechanism allows DNA to be transferred from one bacterium to another. Credit: HHU / Khaled Selim
Understanding natural competence
“c-di-AMP belongs to one the relatively newly discovered class of ‘cyclic dinucleotide-type second messengers,’ whose cellular functions are not yet fully understood,” said Professor Selim. “In these studies, we show that the ComFB proteins sense c-di-AMP and additionally c-di-GMP and are essential for regulating the natural competence and bacterial motility.”
‘Natural competence’ refers to the ability of bacterial cells to uptake DNA molecules and integrate them into their own genome – facilitating genetic exchange. This process is a major component in the spread of antibiotic resistance from a few bacteria to entire populations – and even across species boundaries.
Implications for fighting antibiotic resistance
While the mechanism has been confirmed in the bacteria studied, researchers say it is still unclear which other bacterial groups may use it.
“If pathogenic bacteria with clinical relevance also use it, this could pave the way for new strategies to fight multi-resistant bacteria,” said Professor Salim.
Related topics
Analysis, Antibiotics, Antimicrobials, Biofilms, DNA, Drug Targets, Microbiology, Translational Science
Related conditions
antibiotic resistance