CRISPR activation screening: finding genes that interact with SARS-CoV-2
Posted: 17 January 2022 | Dr Greg Neely (University of Sydney) | No comments yet
Dr Greg Neely, University of Sydney, explains how he and his team used pooled whole genome CRISPR activation screening to identify LRRC15 as a SARS-CoV-2 spike-interacting protein.


Can you briefly outline what whole genome CRISPR screening is?
CRISPR is a highly efficient and simple way to change the code of DNA. We can use CRISPR to edit DNA so that the gene does not code for a protein anymore. As it is so efficient, we can actually target most of the human genome with this technology.
Most importantly, it is scalable, meaning that instead of studying the role of one gene in a process, we can now use CRISPR to target every human gene. Of course, if you turned off every human gene, it would kill a human cell, so instead we can grow millions of human cells using a cell line and then turn a single gene off in each of the cells within the mixture. We can then add something else, such as SARS-CoV-2, where the coronavirus would kill all the unedited cells in this experiment and also kill cells that have a gene turned off that the coronavirus does not need.
However, if a cell is missing something that the coronavirus needs to enter or replicate or kill our cells, then those cells will become resistant to the virus and survive when the experiment is over. We then use DNA sequencing to identify the genes that, when targeted, make a cell resistant to the virus. This is essentially what a whole genome CRISPR screen is; like finding a needle in a haystack, but with a magnet.
How do you utilise whole genome CRISPR screening in your research?
The approach we use was developed by others; the logic was laid down by yeast genetics researchers and brought to the human system by people like Thijn Brummelkamp in the Netherlands. While the first CRISPR knockout (KO) library was made by Feng Zhang at the Broad Institute, we currently use the TKO3 library made by Jason Moffatt in Toronto. Our contribution is applying this technology to new areas, like venom research, or looking for multidrug resistance genes. We have also applied it to our new study where instead of looking at cell death, we used CRISPR to look for genes that could bind the SARS-CoV-2 spike protein.
The approach we used is standard in the world of CRISPR screening. After we identify the genes that all have a similar phenotype, such as resistance to a venom, we then use bioinformatics to try and figure out if this list of genes has anything in common that was not obvious. If a group of genes are all involved in one process – like “cholesterol biosynthesis”, which is what we found when we screened box jellyfish venom – then we can use this information to target cholesterol and stop the damage, which worked for this venom.
Can you describe a recent study where you used a CRISPR screen?
One of our most interesting unpublished projects, currently in preprint in bioRxiv, involved using CRISPR activation screening to find new human genes that promote cell interaction with SARS-CoV-2 (Figure 1). In this case, instead of turning off genes (a KO screen), we turned genes on. We used a modified version of CRISPR where the enzyme that usually cuts DNA (Cas9) no longer has the ability to cut; instead we linked transcriptional activators to Cas9. Then, when the guide RNA brings the dead Cas9 to the target gene, it recruits transcriptional machinery that turns on that gene. This is called “CRISPR activation” and using this we can identify human genes that have a specific positive activity, versus looking for loss of this activity. Rather than looking for drug targets, this process can teach us which human genes have a desired activity, making these genes lead molecular targets for a drug.
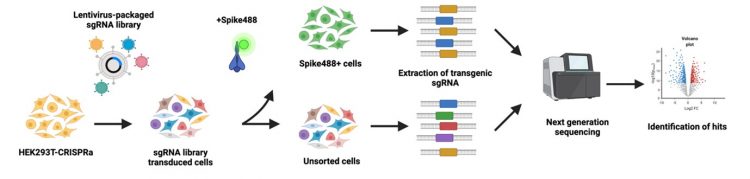

Figure 1: General schematic for how a pooled CRISPR screen works to identify enrichment of a novel cell phenotype, in this case the ability to bind to SARS-CoV-2 spike protein [created with BioRender.com].
Using CRISPR activation, we took human cells that cannot interact with SARS-CoV-2, activated all their genes in cell mixtures and then used flow cytometry to pull out genes that are either SARS-CoV-2 receptors themselves or are master regulators upstream of coronavirus receptors. By doing this, we found a new receptor called leucine rich repeat protein 15 (LRRC15) that we think acts as a new antiviral innate immune barrier. It is related to toll like receptors and when we add this gene to cells, they can then interact with the SARS-CoV-2 spike protein. We found this receptor cannot support infection, but instead seems to collect soluble virus and sequester it, so it cannot infect its intended target. By researching the mechanisms involved in how this gene acts in more detail, we found LRRC15 is expressed on lung fibroblasts, the cells that help repair the lung after injury but that also result in lung fibrosis, a major aspect of “long COVID”. From this screen where we went in knowing almost nothing, we identified this critical new host defence protein. Putting the data pieces together, we can then start to better understand the puzzle of how long COVID works. We can start to reformulate or target LRRC15 and this could then potentially be the basis for new long COVID drugs or even general antifibrosis drugs.
What challenges did you face during this research?
The hardest challenge currently is in moving CRISPR screening beyond merely cell death screens. We have overcome this by designing new highly specific screening readouts that are amenable to high-throughput screening. For example, sorting cells that have acquired a desired phenotype using flow cytometry or using transgenic reports that lead to a selectable outcome, like expression of green fluorescence protein or expression of an antibiotic selection marker. In my opinion, these new, yet more complicated, non-lethal screens will be one major future for drug target discovery.
ARTICLE: CRISPR screens and novel drug targets…
Dr Jeffrey Rathmell and Ayaka Sugiura from Vanderbilt University discuss why inhibiting or genetically deleting the MTHFD2 enzyme using CRISPR reduced disease severity in multiple inflammatory disease models.
How do you think the use of CRISPR in this context will evolve in the future?
As pooled CRISPR screening becomes more common place, it will be used in all aspects of basic and applied medical research. Instead of, or in combination with, running RNA sequencing experiments, researchers will run either whole genome or targeted CRISPR screens to identify novel mechanisms.
For the biotech and pharmaceutical industry, one of the easiest applications of pooled CRISPR screening is to characterise exactly how a drug acts. As long as the drug does something that we can detect in human cells, we can then screen for loss of this action and whatever genes we hit are therefore involved either directly or indirectly in the drug mechanism, which we can then sort out quickly with standard biochemistry.
One of our future interests is using CRISPR activation to modulate stem cells and turn them into more accurate versions of adult human tissues, then in that way we can start to build effective cell or tissue transplants and replace body parts with our own stem cell-derived materials as we age. While I think this is very possible within the next 20 years, first we need to get the most basic stem cell therapies approved for conditions like Parkinson’s or type 1 diabetes.
Dr Greg Neely completed his PhD in human immunology (AIDS related pathogens) at the University of Calgary, Canada and went on to train in functional genomics at the IMBA in Vienna, Austria. Since 2011 he has been running a lab at the University of Sydney, Australia using functional genomics approaches to find novel human disease genes and pathways. His lab is using flies, mice and human stem cells combined with CRISPR screening and single cell sequencing to identify new human genes and genetic pathways controlling human disease. His primary focus is on pain biology, but since 2020 has been applying functional CRISPR screening to study SARS-CoV-2 biology.
Related topics
CRISPR, Flow Cytometry, Genetic Analysis, Genome Editing, Molecular Targets, Protein, Proteomics
Related conditions
Covid-19, Long COVID, Lung fibrosis, Parkinson's disease, type 1 diabetes
Related organisations
Broad Institute
Related people
Dr Feng Zhang, Jason Moffatt, Thijn Brummelkamp