Mechanism-informed phenotypic screening – the missing link for cancer drug discovery?
Posted: 8 June 2016 | David S. Bailey (IOTA Pharmaceuticals), Sheraz Gul (Fraunhofer-IME SP) | No comments yet
Cancer cells exhibit huge phenotypic diversity, with many well-defined phenotypes, such as invasion, deregulated proliferation and immune evasion, contributing to disease outcomes, making them uniquely amenable to phenotypic screening1…

Our ability to study and analyse the functional characteristics of these phenotypes has been greatly enhanced by genetics and genomics, both at the patient stratification and malignant cell levels. Phenotypic screening in general, and in cancer specifically, has benefitted from an increasing understanding of the relationship between genotype and phenotype, informing the basis of current approaches to both phenotypic and target-based drug discovery across the oncology spectrum (summarised in Figure 1).
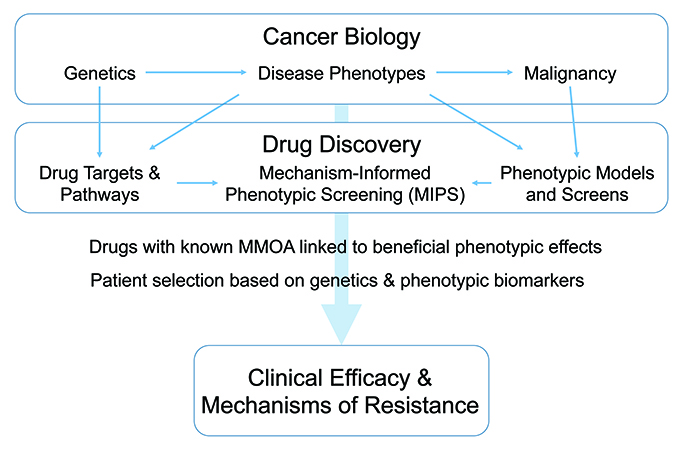

Figure 1: Phenotypic screening in cancer.
Glioblastoma – a rare cancer
Although genomics has provided a mass of molecular information to investigate the aetiology of cancer, most cancers still lack adequate long-term, sustainable therapy. This is especially the case with rare cancers, which are characterised by small and dispersed patient populations, highly specialised and often novel treatment regimens (requiring the engagement of regulatory authorities during development), and ultimately a limited market potential with even the best therapies providing relatively low commercial returns.
An example of such a rare cancer is glioblastoma multiforme (GBM), a phenotypically complex and challenging disease to treat2. Nevertheless, recent advances in GBM research discussed in this short article give us considerable hope of applying new phenotypic screening methods for patient diagnosis, follow-up and treatment evaluation, revealing therapeutic options so far missed in treating this challenging rare disease.
Advancing the development of new therapeutic options
At present there are no effective therapies for patients suffering from GBM. In this context, there is no doubt that the availability of preclinical diagnostic information for individual patients and their tumours would provide benefit both to the patient (through precise phenotyping of their form of the disease thereby eventually helping to administer appropriate therapies), and to the clinician who will be able to compare and contrast response rates using particular drug regimens in defined clinical GBM phenotypes. The distress caused by current treatment options with limited efficacy (namely temozolomide and radiotherapy) is evident, especially in younger patients who find such treatments daunting at best. Even marginal improvements in longevity and quality of life will represent a significant benefit to patients with this disease.
Drug repurposing


Drugs repurposing could shed light on new therapies for GBM
One route for searching out effective therapies in GBM as a prelude to new drug discovery may be repurposing. There is hope that drug discovery approaches to GBM may be accelerated using known bioactive compounds, including drugs already registered at the EMA and FDA with known and specific modes of action, which are immediately available for clinical evaluation3. Also, it is evident from the widespread appearance of GBM as a secondary clinical target that many experimental therapies being developed for use in cancers with larger markets are being trialled for GBM in the clinic.
The availability of patient-centric preclinical diagnostics to measure efficacy in GBM may also encourage companies to see GBM as a sound investment in personalised medicine. In addition, compounds active in GBM preclinical models could well translate by molecular mechanism into other cancers, giving access to larger therapeutic markets. The translation of molecular mechanisms as therapeutic approaches to treating disparate diseases with commonalities in cellular phenotypes is the basis of the concept of mechanism-informed phenotypic screening (MIPS1).
Surgical intervention and phenotypic screening
A suite of new surgical methods to interrogate GBM phenotypic heterogeneity have been reported4. Specific cell-based systems as research tools to evaluate disease progression and study the functional behaviour of GBM cells isolated from patient tumours have also been developed5,6, through which to facilitate GBM drug discovery7,8. The availability of such tools makes early use of phenotypic screening in GBM a realistic preclinical option.
Targeting stem cells and their proteins
Brain tumour stem cells (BTSCs) have been implicated in GBM as initiators of disease, and potentially in causing resistance to therapy. A recent study using a novel in vitro GBM model indicated that chemoradiotherapy treatment might actually increase stem cell gene expression and GBM self-renewal, and has therefore been suggested as a drug discovery target for GBM9. As an hypoxic microenvironment has been reported to play a role in controlling GBM BTSCs, understanding the mechanism of action of the microenvironment and its interplay with different cell types within the tumour mass – and the therapeutic implications resulting from this – may also be highly relevant to GBM progression10.

X-ray crystallography is a powerful target-based technology in drug discovery.
Several individual molecular targets (for example, the STAT3, PLK-1 and the CDK genes, CDK4 and CDK6) have been suggested as critical mediators of GBM tumour growth and therapeutic resistance. A small molecule, non-phosphorylated-STAT3 inhibitor that binds to STAT3 protein has been identified (KD = 300nM) that kills GBM BTSCs and suppresses STAT3 phosphorylation and its downstream transcriptional targets11. GBM is also acutely susceptible to proliferative disruption by Plk1 inhibitors – both on their own12 and in combination with BRAF inhibitors13 – while dual inhibitors of CDK4 and CDK6 have also been shown to increase survival time in rat orthotopic U87MG xenograft models14. These advances hold hope for the development of a new echelon of GBM drugs.
Making an impact in GBM drug discovery and advancing beyond state-of-the-art
Current approaches to drug discovery are leveraged by powerful target-based technologies such as high-throughput sequencing and X-ray crystallography. However, there are many diseases in which molecular targets are not enough. What is required is a deep knowledge of functional biology. GBM is one of these diseases.
From a clinical perspective, GBM is phenotypically diverse, therapeutically recalcitrant and highly aggressive. These characteristics reflect a highly complex underlying functional biology. We need to know much more about the behaviour of individual cell lineages in this rare and intractable disease. As a further element of such programmes, we would anticipate the use of novel non-invasive techniques (for example, high-resolution magnetic resonance imaging and serum diagnostics, such as circulating free DNA profiling) to explore the application of longitudinal diagnostics within existing and proposed GBM clinical studies.
Recent research15 has indicated that GBM evolves over time, so it is important to define the exact clinical stage (GBM phenotype) of patients presenting in the clinic. Currently the diagnostic tools to do this are weak, and there is a pressing need to obtain new and reliable diagnostic markers, bringing powerful genetic and phenotypic approaches to bear on patient derived material, such as primary cell cultures5 and liquid biopsies16. These diagnostics may have commercial value, but above all will enable comparison of the cellular phenotypes evolving in culture with the clinical progress of disease in individual patients.
We see powerful synergies between cutting-edge cell biology, exemplified by in-depth phenotypic screening, and new patient-centric GBM diagnostics. Defining the staging and phenotypic variation between individual GBM patients may enable the clinician to place patients in treatment regimens experimentally validated for particular GBM phenotypes.
GBM – the way forward?
With the challenges associated with GBM drug discovery, the following tools and technologies should be prioritised to address this:
- Developing patient-specific cell libraries that are genetically and clinically annotated from which to develop cell-based approaches to new drug discovery in a personalised, patient-centric context5,6
- Establishing proof of concept for an integrated phenotypic screening and target discovery platform, using GBM as an exemplar rare disease where there are both adequate pre-clinical patient-centric models and significant unmet clinical needs. We see MIPS providing a basis for this1
- Developing methodologies and protocols, and validating them using patient-derived cell platforms with further dissemination into academic and pharma sectors through existing and new partnerships17,18
- And finally, as a strategic outcome, fostering close academic/commercial collaborations to establish lab-to-clinic pipelines with a clear trajectory towards efficient patient delivery of candidate therapies19. The establishment of global networks to facilitate the development of GBM therapeutics is a high priority.
Conclusions
Current approaches to drug discovery are leveraged by powerful target-based technologies such as high-throughput sequencing and X-ray crystallography. However, there are many diseases in which addressing individual molecular targets is simply not enough. What is required is a deep knowledge of functional biology: GBM is one of these diseases.
From a clinical perspective GBM is phenotypically diverse, therapeutically recalcitrant and highly aggressive. These characteristics reflect a highly complex underlying functional biology: there is a need to understand much more about the behaviour of individual cell lineages in this rare and intractable disease. This requires a multipronged approach to bring together clinical excellence in GBM management, cutting-edge cell biology exemplified by powerful phenotypic screening technologies, and informative patient-centric diagnostics, through which to identify clinical candidates with potential in GBM. Powerful synergies between patient-centric clinical diagnostics and lab-based phenotypic drug discovery techniques, such as MIPS, make the translation of therapies between GBM and more prevalent cancers highly likely.
Biography




References
- Moffat JG, Rudolph J, Bailey D. Phenotypic screening in cancer drug discovery – past, present and future. Nat Rev Drug Discov. 2014 Aug;13(8):588-602
- Watts C, Price SJ, Santarius T. Current concepts in the surgical management of glioma patients. Clin Oncol (R Coll Radiol). 2014 Jul;26(7):385-94
- Jiang P, Mukthavaram R, Chao Y, Bharati IS, Fogal V, Pastorino S, Cong X, Nomura N, Gallagher M, Abbasi T, Vali S, Pingle SC, Makale M, Kesari S. Novel anti-glioblastoma agents and therapeutic combinations identified from a collection of FDA approved drugs. J Transl Med.2014 Jan 17;12:13
- Piccirillo SG, Dietz S, Madhu B, Griffiths J, Price SJ, Collins VP, Watts C. Fluorescence-guided surgical sampling of glioblastoma identifies phenotypically distinct tumour-initiating cell populations in the tumour mass and margin. Br J Cancer. 2012 Jul 24;107(3):462-8
- Fael Al-Mayhani TM1, Ball SL, Zhao JW, Fawcett J, Ichimura K, Collins PV, Watts C. An efficient method for derivation and propagation of glioblastoma cell lines that conserves the molecular profile of their original tumours. J Neurosci Methods. 2009 Jan 30;176(2):192-9
- Al-Mayhani MT, Grenfell R, Narita M, Piccirillo S, Kenney-Herbert E, Fawcett JW, Collins VP, Ichimura K, Watts C. NG2 expression in glioblastoma identifies an actively proliferating population with an aggressive molecular signature. Neuro Oncol. 2011 Aug;13(8):830-45
- Kenney-Herbert EM, Ball SL, Al-Mayhani TM, Watts C. Glioblastoma cell lines derived under serum-free conditions can be used as an in vitro model system to evaluate therapeutic response. Cancer Lett. 2011 Jun 1;305(1):50-7
- Setua S, Ouberai M, Piccirillo SG, Watts C, Welland M. Cisplatin-tethered gold nanospheres for multimodal chemo-radiotherapy of glioblastoma. Nanoscale. 2014 Sep 21;6(18):10865-73
- Qazi MA, Vora P, Venugopal C, McFarlane N, Subapanditha MK, Murty NK, Hassell JA, Hallett RM, Singh SK. A novel stem cell culture model of recurrent glioblastoma. J Neurooncol. 2016 Jan;126(1):57-67
- Persano L, Rampazzo E, Basso G, Viola G. Glioblastoma cancer stem cells: role of the microenvironment and therapeutic targeting. Biochem Pharmacol. 2013 Mar 1;85(5):612-22
- Haftchenary S, Luchman HA, Jouk AO, Veloso AJ, Page BD, Cheng XR, Dawson SS, Grinshtein N, Shahani VM, Kerman K, Kaplan DR, Griffin C, Aman AM, Al-Awar R, Weiss S, Gunning PT. Potent Targeting of the STAT3 Protein in Brain Cancer Stem Cells: A Promising Route for Treating Glioblastoma. ACS Med Chem Lett. 2013 Sep 8;4(11):1102-7
- Danovi D, Folarin A, Gogolok S, Ender C, Elbatsh AM, Engström PG, Stricker SH, Gagrica S, Georgian A, Yu D, U KP, Harvey KJ, Ferretti P, Paddison PJ, Preston JE, Abbott NJ, Bertone P, Smith A, Pollard SM. A high-content small molecule screen identifies sensitivity of glioblastoma stem cells to inhibition of polo-like kinase 1. PLoS One. 2013 Oct 30;8(10):e77053
- Lerner RG, Grossauer S, Kadkhodaei B, Meyers I, Sidorov M, Koeck K, Hashizume R, Ozawa T, Phillips JJ, Berger MS, Nicolaides T, James CD, Petritsch CK. Targeting a Plk1-controlled polarity checkpoint in therapy-resistant glioblastoma-propagating c Cancer Res. 2015 Dec 15;75(24):5355-66
- Raub TJ, Wishart GN, Kulanthaivel P, Staton BA, Ajamie RT, Sawada GA, Gelbert LM, Shannon HE, Sanchez-Martinez C, De Dios A. Brain Exposure of Two Selective Dual CDK4 and CDK6 Inhibitors and the Antitumor Activity of CDK4 and CDK6 Inhibition in Combination with Temozolomide in an Intracranial Glioblastoma Xenograft. Drug Metab Dispos. 2015 Sep;43(9):1360-71
- Sottoriva A, Spiteri I, Piccirillo SG, Touloumis A, Collins VP, Marioni JC, Curtis C, Watts C, Tavaré S. Intratumor heterogeneity in human glioblastoma reflects cancer evolutionary dynamics. Proc Natl Acad Sci U S A. 2013 Mar 5;110(10):4009-14
- Santiago-Dieppa DR, Steinberg J, Gonda D, Cheung VJ, Carter BS, Chen CC. Extracellular vesicles as a platform for ‘liquid biopsy’ in glioblastoma patients. Expert Rev Mol Diagn. 2014 Sep;14(7):819-25
- Wehmas LC, Tanguay RL, Punnoose A, Greenwood JA. Developing a Novel Embryo-Larval Zebrafish Xenograft Assay to Prioritize Human Glioblastoma Therapeutics. Zebrafish. 2016 May 9. [Epub ahead of print]
- Fan Y, Nguyen DT, Akay Y, Xu F, Akay M. Engineering a Brain Cancer Chip for High-throughput Drug Screening. Sci Rep. 2016 May 6;6:25062
- Guilfoyle MR, Weerakkody RA, Oswal A, Oberg I, Jeffery C, Haynes K, Kullar PJ, Greenberg D, Jefferies SJ, Harris F, Price SJ, Thomson S, Watts C. Implementation of neuro-oncology service reconfiguration in accordance with NICE guidance provides enhanced clinical care for patients with glioblastoma multiforme. Br J Cancer. 2011 Jun 7;104(12):1810-5
Related topics
Biologics, Drug Discovery, Genomics, Oncology, Protein, Screening, Therapeutics
Related conditions
Glioblastoma