Human pluripotent stem cell-derived cardiomyocytes in drug discovery and development
Posted: 12 June 2015 | Arne Hansen (University Medical Center Hamburg-Eppendorf and DZHK), Boris Greber (Max Planck Institute for Molecular Biomedicine and Chemical Genomics Centre of the Max Planck Society), Dennis Schade (Technical University of Dortmund) | No comments yet
Procedures for cardiac differentiation of human pluripotent stem cells (hPSCs) have greatly improved over the past few years. The facilitated derivation of hPSC-cardiomyocytes has enabled the thorough assessment of their physiology and manipulation of their properties. Moreover, patient-specific hPSCs generated through cellular reprogramming allow for cardiac disease modelling, discovery of underlying disease mechanisms and drug evaluation…
This review highlights the utility of small molecules in promoting differentiation of hPSCs into cardiomyocytes, in stimulating their specification and maturation in vitro, as well as in patient-specific drug assessment and screening. We discuss principles and recent developments of directed cardiac induction, the maturation status of hPSC-cardiomyocytes as compared to in vivo standards, as well as emerging in vitro applications.
Cardiac differentiation of hPSCs
Human embryonic stem cells and human induced pluripotent stem cells (collectively: hPSCs) can differentiate along all three germ layers to give rise to various somatic cell types in vitro, including cardiomyocytes (CMs). Procedures for inducing CM differentiation of hPSCs have greatly improved over the past few years. Earlier protocols were largely based on spontaneous differentiation using permissive rather than inductive culture conditions. To some degree, these protocols also relied on the intrinsic propensity of certain hPSC lines to differentiate along the cardiac lineage by default. Nonetheless, important insights were gained from these efforts, such as the finding that insulin – a commonly used media additive – strongly inhibits human cardiac induction1.
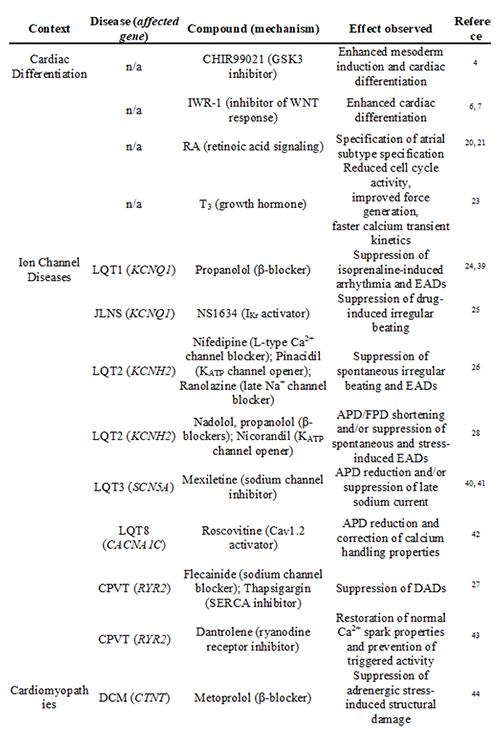
Table 1: Beneficial effects of tool compounds and drugs used in hPSC-based differentiation and disease modelling. Key: LQT: long QT syndrome, EADs: early afterdepolarisations, JLNS: Jervell and Lange-Nielsen syndrome, APD: action potential duration, FPD: field potential duration, CPVT: catecholaminergic polymorphic ventricular tachycardia, DADs: delayed afterdepolarisations, DCM: dilated cardiomyopathy, HCM: hypertrophic cardiomyopathy, DMCM: diabetes mellitus cardiomyopathy, BNP: brain natriuretic peptide.
A paradigm shift then led to the development of directed differentiation procedures which tend to mimic signalling mechanisms orchestrating embryonic development2. The use of small molecules as ‘chemical tool compounds’ has greatly improved this approach. Directed cardiac differentiation protocols are commonly based on an initial stimulation of mesoderm-inducing signalling pathways, notably bone morphogenetic proteins (BMP) and the protein ligands WNTs, followed by a cardiac specification phase driven by WNT inhibition. Moreover, beyond these biphasic signalling requirements, cardiac induction necessitates high cell densities, which can either be achieved through formation of three-dimensional aggregates (embryoid bodies; EBs) or under confluent monolayer culture conditions. Under such conditions, however, recombinant protein factors, such as BMP4 or WNT3A, may have limited access to the cells. For this and other reasons such as cost-effectiveness, small molecules have largely replaced the use of recombinant factors in cardiac differentiation protocols wherever possible. For example, the GSK3β inhibitor CHIR99021 has replaced WNT3A as a surrogate for activating canonical WNT signalling, and distinct small molecule WNT inhibitors (e.g., the porcupine inhibitor C593) have replaced the use of recombinant DKK14-6,7 (Table 1).
As much as the defined stage-wise treatment of hPSCs with these molecules matters, so does the state in which hPSCs reside prior to initiating differentiation. For example, significant spontaneous differentiation in hPSC cultures will inevitably compromise subsequent cardiac induction. Hence, the development of efficient CM differentiation protocols has also been advanced by the improvement of hPSC culture systems. Contemporary culture systems are based on chemically-defined media formulations, some of which make use of small molecule inhibitors to suppress spontaneous differentiation8, 9. Furthermore, periodic single-cell splitting of hPSCs, as opposed to colony culture, has been shown to reduce heterogeneity and enhance subsequent cardiac differentiation efficiencies10, 11. Single cell passaging had previously been hampered by the notorious propensity of dissociated hPSCs to undergo apoptosis but the small molecule Rho-kinase inhibitor Y-27632 was found to overcome this limitation12.
Upon combining these strategies, hPSCs can now be converted into CM-like cells at efficiencies above 80%3,11,13. Interestingly, these procedures tend to overcome inter-line variability in that they are applicable to independent hPSC lines. These improvements have been crucial in the field of cardiac disease modelling using patient-specific induced pluripotent stem cells (hiPSCs), where researchers are bound to working with specific lines of a given genetic background. The success of using directed differentiation protocols confirms the overall idea of guiding hPSCs through defined intermediate developmental stages in a controlled manner. However, it is crucial with these procedures to strictly control the undifferentiated state of the hPSCs, cell densities, as well as the activities of added factors (unpublished observations). Further improving the robustness of directed cardiac differentiation protocols should therefore be given a priority in future investigations.
Properties of hPSC-derived cardiomyocytes
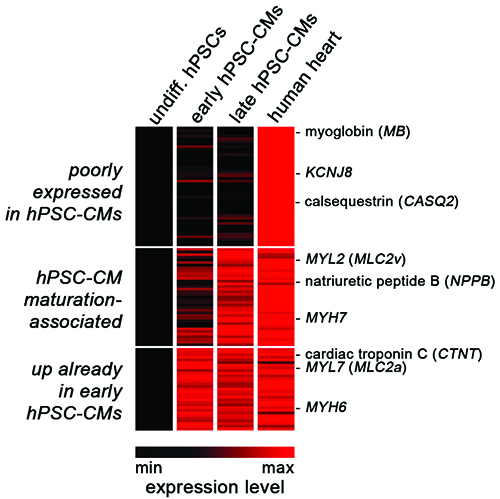
Figure 1: Shared and divergent gene expression in hPSC-CMs and human heart. Heat map representation of normalised profiling data comparing undifferentiated hPSCs, early hPSC-CMs (one week old), maturated hPSC-CMs (five weeks old), and human heart tissue. Three classes of cardiac genes were defined according to their expression patterns (see labelling on the left).
CM formation from hPSCs becomes readily apparent through spontaneous cellular contraction, as early as approximately one week after differentiation. hPSC-CMs can be maintained in culture for many weeks thereafter, and their regular beating activity implies that basic structural and electrophysiological properties must be in place. Indeed, even early hPSC-CMs express a number of cardiac-specific genes such as CTNT (cardiac troponin), MYH6 (myosin heavy chain), and CACNA1C (L-type calcium channel), at levels comparable to those in human heart tissue (Figure 1, bottom panel11).
Their rapid induction seems to be in agreement with early human development, where the beating primitive heart tube is the first organ to form. However, it is widely accepted – and unsurprising – that early hPSC-CMs display overall immature structural and physiological features. Structurally, hPSC-CMs tend to be irregularly shaped, do not show a well-aligned sarcomeric structure, and lack T-tubles14. Moreover, hPSC-CMs express only low levels of CASQ2 (calsequestrin), although they do store Ca2+ intracellularly15,16,17 (Figure 1, upper panel). Electrophysiologically, hPSC-CMs show a more depolarised resting membrane potential, slower upstroke kinetics, and slow depolarisation during the late phase of the action potential, as compared to adult cardiomyocytes16. These features may also explain their persistent automaticity in culture.
Nonetheless, hPSC-CMs functionally express a number of key ion channels involved in action potential generation, and ventricular, atrial, and pacemaker-like cells identified in these cultures have been extensively characterised16,18. Moreover, it has been realised that prolonged in vitro culture of hPSC-CMs promotes their maturation in a number or respects. For instance, calcium transient amplitudes, sarcomeric organisation, and the expression of several ion channel-encoding genes like KCNQ1 gradually increase over time11,19. These, as well as several structural maturation markers like MYH7 and ventricular-specific MYL2, tend to reach human heart-like expression levels after several weeks in culture (Figure 1, middle panel11). The high expression levels of MYL2 in late hPSC-CMs may also reflect the fact that the majority of hPSC-CMs commonly acquire a ventricular identity by default. Interestingly, the small signalling molecule retinoic acid has recently been shown to redirect terminal cell fate to an atrial identity, suggesting that CM subtype-specific differentiation may now be possible on demand20,21 (Table 1).
Furthermore, several strategies are being explored to accelerate and improve maturation of hPSC-CMs. These approaches include 3D tissue engineering, mechanical stretching, electrical stimulation, co-culture with non-cardiomyocytes, and genetic manipulation (reviewed in 15). For example, forced expression of KCNJ2 (encoding the inwardly-rectifying potassium channel Kir2.1) has been shown to confer a more adult-like electrophysiological phenotype to hPSC-CMs22. In addition, a particularly simple and non-invasive strategy presents administration of the physiological growth hormone triiodothyronine (T3). Addition of this small compound to the culture medium was shown to promote maturation by reducing cell cycle activity, increasing calcium transient kinetics, and improving contractile force generation23 (Table 1). Despite these improvements, it shall be noted that even in late hPSC-CMs, a number of cardiac genes are essentially not expressed (Figure 1, top panel). Conversely, some of the immature features of hPSC-CMs may also be beneficial for downstream applications, that is, erasing them could also be counterproductive. For example, spontaneous beating of hPSC-CMs may conveniently be exploited in assessing proarrhythmic effects of drugs, although silent CMs could also be stimulated through electrical pacing.
Applications based on hPSC-CMs
The methodology of generating hiPSCs through direct reprogramming of patient cells, together with the availability of improved cardiac differentiation protocols, is appreciably transforming the field of cardiac disease modelling and drug testing. Despite the aforementioned shortcomings, the great advantage that the system offers is a human cardiac cell context. In principle, one can distinguish between approaches to identify drugs or targets that induce a desired (disease-correcting) effect or, alternatively, to reveal adverse cardiovascular side-effects of drugs (cardiotoxicity; Figure 2).
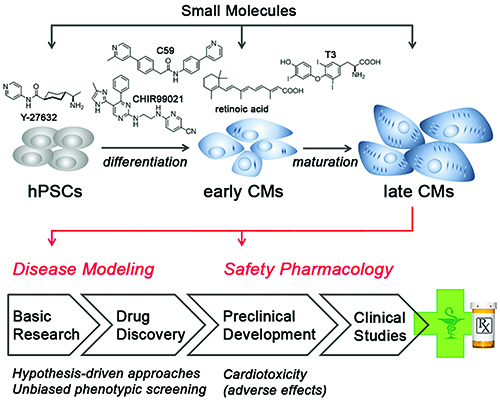
Figure 2: Applications enabled by hPSC-derived cardiomyocytes. Small molecules are used as chemical tools in hPSC maintenance and differentiation (top). Later-stage hPSC-CMs are then applied in cardiac disease modelling to investigate pathogenic mechanisms, identify disease-relevant targets, and evaluate specific drugs in a hypothesis-driven manner (bottom). Alternatively, hPSC-CMs may be utilised in unbiased phenotypic screening approaches. At the preclinical stage of drug development, hPSC-CMs may be employed for assessing adverse side-effects of drug candidates. See text for further discussion.
The first cardiac diseases that have been modelled using patient-specific hiPSCs were monogenetic ion channel disorders. The phenotypes associated with this class of diseases are cell autonomous and directly attributable to a given genetic defect. Hence, disease features could directly be inferred from patient-specific hiPSC-CMs, while comparing these to wild-type hiPSC-CMs. Remarkably, most phenotypes revealed in the hiPSC models closely recapitulated disease features seen in the underlying patients, implying that hPSC-CMs indeed present a powerful model. Interestingly, a number of studies have successfully employed drug testing to reverse the disease phenotypes in the cellular models (Table 1). In several reports, these efforts confirmed the beneficial effects of clinically established treatments, for instance, β-blocker therapy in case of long QT syndrome 124. In other cases, drug evaluation was based on more hypothesis-driven approaches targeting ion channels other than the mutated one. This not only balanced the primary electrophysiological consequences of the underlying mutation but it also protected patient hiPSC-CMs from rather global adverse responses, such as irregular beating25-27,28 (Table 1). In several reports, hiPSC-CM models also served to decipher underlying disease mechanisms. In a hypertrophic cardiomyopathy study, Lan et al. found that a missense mutation in the structural MYH7 gene caused hypertrophy at the single-cell level, which could be balanced by restoring Ca2+ homeostasis29. Collectively, these reports provide compelling support for the utility of patient-specific hiPSCs for cardiac disease modelling, discovery of disease mechanisms and drug evaluation.
In future investigations, it will be interesting to also employ patient-specific hiPSC-CMs in more non-hypothesis-driven approaches for candidate drug identification30. Classically, novel drug candidates are discovered from target-centric approaches, where mostly, a biochemical assay is used to screen for hits in large small molecule libraries. By contrast, phenotypic screening approaches based on hiPSC-CMs could more faithfully reflect the overall pathophysiological situation and also reveal unexpected, potentially novel drug targets. In fact, drug screening-compatible phenotypic assays have already begun to be employed in this area31,32. For instance, Drawnel et al. used hPSC-derived CMs to model diabetic cardiomyopathy32. Interestingly, features of this multifactorial disease such as structural and functional disarray could readily be induced by applying exogenous stimuli (glucose and corticosteroids). Based on this diabetic cardiomyopathy surrogate, the authors eventually introduced an assay platform to address disease-relevant, screenable endpoints such as the striated sarcomeric pattern of hiPSC-CMs, nuclear area and BNP production. Strikingly, several compounds identified from an unbiased phenotypic screen on wild-type cells also rescued disease phenotypes in patient-specific CMs (32; Table 1).
Moreover, hPSC-CMs are increasingly being explored and used for safety pharmacology and toxicology applications in early, preclinical drug discovery. Cardiovascular toxicity most frequently accounts for adverse drug effects and represents a major cause for drug attrition in late-stage clinical development, or even withdrawal of marketed drugs33,34. Due to shortcomings of existing assays based on heterologous expression systems or animal trials, there is a pressing need to more reliably reveal such risks and avoid false positive outcomes. High-quality cardiotoxicity prediction will help in the decision-making progress on whether or not to continue with a specific drug candidate, ideally before entering clinical trials. More predictive tools would therefore benefit patients but also reduce costs in the drug development process. hPSC-CMs may present a powerful system in this context and hence, several readout systems based on their physiology are currently being explored. These test systems are based on integrated readouts of complex parameters, including field potential prolongation on multielectrode arrays, analysis of contractile force, or drug-induced irregular beating35-37. These phenotypic assays could well complement the in-depth electrophysiological characterisation in heterologous expression systems38, whereas the most suited platforms and assay standards remain to be defined.
Acknowledgements: The authors wish to acknowledge support from the Chemical Genomics Centre of the Max Planck Society and the German Federal Ministry of Education and Research (BMBF, grant 131605).
Biographies
 DR. DENNIS SCHADE is a pharmacist by training and received his PhD in medicinal chemistry at the Christian-Albrechts-University of Kiel. He stayed there for a short time before undertaking postdoctoral studies as a DFG Research Fellow at the Sanford-Burnham Medical Research Institute and the Human BioMolecular Research Institute (San Diego). Funded by the German Federal Ministry of Education and Research (BMBF), he became ‘Group Leader Medicinal Chemistry’ at the Technical University Dortmund (10/2012) for independent research in the field of small molecule stem cell modulators. His current research focus is on the development of ‘Chemical Tools for Cardiac Regeneration’ with a general interest in drug design and development, cardiovascular diseases, stem cells and regenerative medicine, phenotypic assays and pharmacokinetics.
DR. DENNIS SCHADE is a pharmacist by training and received his PhD in medicinal chemistry at the Christian-Albrechts-University of Kiel. He stayed there for a short time before undertaking postdoctoral studies as a DFG Research Fellow at the Sanford-Burnham Medical Research Institute and the Human BioMolecular Research Institute (San Diego). Funded by the German Federal Ministry of Education and Research (BMBF), he became ‘Group Leader Medicinal Chemistry’ at the Technical University Dortmund (10/2012) for independent research in the field of small molecule stem cell modulators. His current research focus is on the development of ‘Chemical Tools for Cardiac Regeneration’ with a general interest in drug design and development, cardiovascular diseases, stem cells and regenerative medicine, phenotypic assays and pharmacokinetics.
 PROFESSOR DR. ARNE HANSEN has a degree in medicine from the University of Hamburg and is a board-certified expert in experimental and clinical pharmacology. He is leading the cardiac tissue engineering group at the Department of Experimental Pharmacology and Toxicology, University Medical Center Hamburg-Eppendorf. He is involved in projects related to in vitro human cardiac tissue models.
PROFESSOR DR. ARNE HANSEN has a degree in medicine from the University of Hamburg and is a board-certified expert in experimental and clinical pharmacology. He is leading the cardiac tissue engineering group at the Department of Experimental Pharmacology and Toxicology, University Medical Center Hamburg-Eppendorf. He is involved in projects related to in vitro human cardiac tissue models.
 DR. BORIS GREBER studied Biochemistry at the University of Potsdam, Germany, and at the John Innes Centre in Norwich, UK. He did his PhD on chemical mutagenesis of mouse ES cells at the Max Planck Institute for Molecular Genetics in Berlin. He then started working with human pluripotent stem cells during his postdoctoral studies with James Adjaye at the same institute and, later, with Hans Schöler at the Max Planck Institute for Molecular Biomedicine in Münster. He is now research group leader affiliated with the MPI Münster as well as with the Chemical Genomics Centre of the Max Planck Society in Dortmund. His research interests are centred around the pluripotency of hPSCs, directed cardiac differentiation, as well as hPSC-based applications like cardiac disease modelling.
DR. BORIS GREBER studied Biochemistry at the University of Potsdam, Germany, and at the John Innes Centre in Norwich, UK. He did his PhD on chemical mutagenesis of mouse ES cells at the Max Planck Institute for Molecular Genetics in Berlin. He then started working with human pluripotent stem cells during his postdoctoral studies with James Adjaye at the same institute and, later, with Hans Schöler at the Max Planck Institute for Molecular Biomedicine in Münster. He is now research group leader affiliated with the MPI Münster as well as with the Chemical Genomics Centre of the Max Planck Society in Dortmund. His research interests are centred around the pluripotency of hPSCs, directed cardiac differentiation, as well as hPSC-based applications like cardiac disease modelling.
References
- Xu XQ, Graichen R, Soo SY, Balakrishnan T, Rahmat SN, Sieh S, et al. Chemically defined medium supporting cardiomyocyte differentiation of human embryonic stem cells. Differentiation. 2008;76(9):958-70
- Murry CE, Keller G. Differentiation of embryonic stem cells to clinically relevant populations: lessons from embryonic development. Cell. 2008;132(4):661-80
- Burridge PW, Matsa E, Shukla P, Lin ZC, Churko JM, Ebert AD, et al. Chemically defined generation of human cardiomyocytes. Nature methods. 2014
- Yamauchi K, Sumi T, Minami I, Otsuji TG, Kawase E, Nakatsuji N, et al. Cardiomyocytes develop from anterior primitive streak cells induced by beta-catenin activation and the blockage of BMP signaling in hESCs. Genes Cells. 2010;15(12):1216-27
- Yang L, Soonpaa MH, Adler ED, Roepke TK, Kattman SJ, Kennedy M, et al. Human cardiovascular progenitor cells develop from a KDR+ embryonic-stem-cell-derived population. Nature. 2008;453(7194):524-8
- Gonzalez R, Lee JW, Schultz PG. Stepwise chemically induced cardiomyocyte specification of human embryonic stem cells. Angewandte Chemie. 2011;50(47):11181-5
- Willems E, Spiering S, Davidovics H, Lanier M, Xia Z, Dawson M, et al. Small-molecule inhibitors of the Wnt pathway potently promote cardiomyocytes from human embryonic stem cell-derived mesoderm. Circulation research. 2011;109(4):360-4
- Chen G, Gulbranson DR, Hou Z, Bolin JM, Ruotti V, Probasco MD, et al. Chemically defined conditions for human iPSC derivation and culture. Nature methods. 2011;8(5):424-9
- Frank S, Zhang M, Scholer HR, Greber B. Small molecule-assisted, line-independent maintenance of human pluripotent stem cells in defined conditions. PloS one. 2012;7(7):e41958
- Burridge PW, Thompson S, Millrod MA, Weinberg S, Yuan X, Peters A, et al. A universal system for highly efficient cardiac differentiation of human induced pluripotent stem cells that eliminates interline variability. PloS one. 2011;6(4):e18293
- Zhang M, Schulte JS, Heinick A, Piccini I, Rao J, Quaranta R, et al. Universal cardiac induction of human pluripotent stem cells in two and three-dimensional formats: implications for in vitro maturation. Stem cells. 2015;33(5):1456-69
- Watanabe K, Ueno M, Kamiya D, Nishiyama A, Matsumura M, Wataya T, et al. A ROCK inhibitor permits survival of dissociated human embryonic stem cells. Nature biotechnology. 2007;25(6):681-6
- Lian X, Hsiao C, Wilson G, Zhu K, Hazeltine LB, Azarin SM, et al. Robust cardiomyocyte differentiation from human pluripotent stem cells via temporal modulation of canonical Wnt signaling. Proceedings of the National Academy of Sciences of the United States of America. 2012;109(27):E1848-57
- Snir M, Kehat I, Gepstein A, Coleman R, Itskovitz-Eldor J, Livne E, et al. Assessment of the ultrastructural and proliferative properties of human embryonic stem cell-derived cardiomyocytes. Am J Physiol Heart Circ Physiol. 2003;285(6):H2355-63
- Yang X, Pabon L, Murry CE. Engineering adolescence: maturation of human pluripotent stem cell-derived cardiomyocytes. Circulation research. 2014;114(3):511-23
- Chen HS, Kim C, Mercola M. Electrophysiological challenges of cell-based myocardial repair. Circulation. 2009;120(24):2496-508
- Hwang HS, Kryshtal DO, Feaster TK, Sanchez-Freire V, Zhang J, Kamp TJ, et al. Comparable Calcium Handling Of Human iPSC-derived Cardiomyocytes Generated By Multiple Laboratories. Journal of molecular and cellular cardiology. 2015
- Ma J, Guo L, Fiene SJ, Anson BD, Thomson JA, Kamp TJ, et al. High purity human-induced pluripotent stem cell-derived cardiomyocytes: electrophysiological properties of action potentials and ionic currents. Am J Physiol Heart Circ Physiol. 2011;301(5):H2006-17
- Sartiani L, Bettiol E, Stillitano F, Mugelli A, Cerbai E, Jaconi ME. Developmental changes in cardiomyocytes differentiated from human embryonic stem cells: a molecular and electrophysiological approach. Stem cells. 2007;25(5):1136-44
- Zhang Q, Jiang J, Han P, Yuan Q, Zhang J, Zhang X, et al. Direct differentiation of atrial and ventricular myocytes from human embryonic stem cells by alternating retinoid signals. Cell research. 2011;21(4):579-87
- Devalla HD, Schwach V, Ford JW, Milnes JT, El-Haou S, Jackson C, et al. Atrial-like cardiomyocytes from human pluripotent stem cells are a robust preclinical model for assessing atrial-selective pharmacology. EMBO molecular medicine. 2015;7(4):394-410
- Lieu DK, Fu JD, Chiamvimonvat N, Tung KC, McNerney GP, Huser T, et al. Mechanism-based facilitated maturation of human pluripotent stem cell-derived cardiomyocytes. Circulation Arrhythmia and electrophysiology. 2013;6(1):191-201
- Yang X, Rodriguez M, Pabon L, Fischer KA, Reinecke H, Regnier M, et al. Tri-iodo-l-thyronine promotes the maturation of human cardiomyocytes-derived from induced pluripotent stem cells. Journal of molecular and cellular cardiology. 2014
- Moretti A, Bellin M, Welling A, Jung CB, Lam JT, Bott-Flugel L, et al. Patient-specific induced pluripotent stem-cell models for long-QT syndrome. N Engl J Med. 2010;363(15):1397-409
- Zhang M, D’Aniello C, Verkerk AO, Wrobel E, Frank S, Ward-van Oostwaard D, et al. Recessive cardiac phenotypes in induced pluripotent stem cell models of Jervell and Lange-Nielsen syndrome: disease mechanisms and pharmacological rescue. Proceedings of the National Academy of Sciences of the United States of America. 2014;111(50):E5383-92
- Itzhaki I, Maizels L, Huber I, Zwi-Dantsis L, Caspi O, Winterstern A, et al. Modelling the long QT syndrome with induced pluripotent stem cells. Nature. 2011;471(7337):225-9
- Itzhaki I, Maizels L, Huber I, Gepstein A, Arbel G, Caspi O, et al. Modeling of catecholaminergic polymorphic ventricular tachycardia with patient-specific human-induced pluripotent stem cells. Journal of the American College of Cardiology. 2012;60(11):990-1000
- Matsa E, Rajamohan D, Dick E, Young L, Mellor I, Staniforth A, et al. Drug evaluation in cardiomyocytes derived from human induced pluripotent stem cells carrying a long QT syndrome type 2 mutation. Eur Heart J. 2011;32(8):952-62
- Lan F, Lee AS, Liang P, Sanchez-Freire V, Nguyen PK, Wang L, et al. Abnormal calcium handling properties underlie familial hypertrophic cardiomyopathy pathology in patient-specific induced pluripotent stem cells. Cell stem cell. 2013;12(1):101-13
- Mercola M, Colas A, Willems E. Induced pluripotent stem cells in cardiovascular drug discovery. Circulation research. 2013;112(3):534-48
- Carlson C, Koonce C, Aoyama N, Einhorn S, Fiene S, Thompson A, et al. Phenotypic screening with human iPS cell-derived cardiomyocytes: HTS-compatible assays for interrogating cardiac hypertrophy. Journal of biomolecular screening. 2013;18(10):1203-11
- Drawnel FM, Boccardo S, Prummer M, Delobel F, Graff A, Weber M, et al. Disease modeling and phenotypic drug screening for diabetic cardiomyopathy using human induced pluripotent stem cells. Cell reports. 2014;9(3):810-21
- Ferri N, Siegl P, Corsini A, Herrmann J, Lerman A, Benghozi R. Drug attrition during pre-clinical and clinical development: understanding and managing drug-induced cardiotoxicity. Pharmacology & therapeutics. 2013;138(3):470-84
- Kola I, Landis J. Can the pharmaceutical industry reduce attrition rates? Nature reviews Drug discovery. 2004;3(8):711-5
- Navarrete EG, Liang P, Lan F, Sanchez-Freire V, Simmons C, Gong T, et al. Screening drug-induced arrhythmia [corrected] using human induced pluripotent stem cell-derived cardiomyocytes and low-impedance microelectrode arrays. Circulation. 2013;128(11 Suppl 1):S3-13.
- Braam SR, Tertoolen L, van de Stolpe A, Meyer T, Passier R, Mummery CL. Prediction of drug-induced cardiotoxicity using human embryonic stem cell-derived cardiomyocytes. Stem Cell Res. 2010;4(2):107-16
- Schaaf S, Shibamiya A, Mewe M, Eder A, Stohr A, Hirt MN, et al. Human engineered heart tissue as a versatile tool in basic research and preclinical toxicology. PloS one. 2011;6(10):e26397
- Kramer J, Obejero-Paz CA, Myatt G, Kuryshev YA, Bruening-Wright A, Verducci JS, et al. MICE models: superior to the HERG model in predicting Torsade de Pointes. Scientific reports. 2013;3:2100
- Egashira T, Yuasa S, Suzuki T, Aizawa Y, Yamakawa H, Matsuhashi T, et al. Disease characterization using LQTS-specific induced pluripotent stem cells. Cardiovasc Res. 2012;95(4):419-29
- Ma D, Wei H, Zhao Y, Lu J, Li G, Sahib NB, et al. Modeling type 3 long QT syndrome with cardiomyocytes derived from patient-specific induced pluripotent stem cells. International journal of cardiology. 2013;168(6):5277-86
- Terrenoire C, Wang K, Tung KW, Chung WK, Pass RH, Lu JT, et al. Induced pluripotent stem cells used to reveal drug actions in a long QT syndrome family with complex genetics. The Journal of general physiology. 2013;141(1):61-72
- Yazawa M, Hsueh B, Jia X, Pasca AM, Bernstein JA, Hallmayer J, et al. Using induced pluripotent stem cells to investigate cardiac phenotypes in Timothy syndrome. Nature. 2011;471(7337):230-4
- Jung CB, Moretti A, Mederos y Schnitzler M, Iop L, Storch U, Bellin M, et al. Dantrolene rescues arrhythmogenic RYR2 defect in a patient-specific stem cell model of catecholaminergic polymorphic ventricular tachycardia. EMBO molecular medicine. 2012;4(3):180-91
- Sun N, Yazawa M, Liu J, Han L, Sanchez-Freire V, Abilez OJ, et al. Patient-specific induced pluripotent stem cells as a model for familial dilated cardiomyopathy. Science translational medicine. 2012;4(130):130ra47
- Huang HP, Chen PH, Hwu WL, Chuang CY, Chien YH, Stone L, et al. Human Pompe disease-induced pluripotent stem cells for pathogenesis modeling, drug testing and disease marker identification. Human molecular genetics. 2011;20(24):4851-64
Related topics
Drug Development, Drug Discovery, In Vivo, Screening, Stem Cells