Open innovation – a collaboration between academia and the pharmaceutical industry to further leverage drug discovery expertise and assets
Posted: 13 December 2019 | Dr David Murray (AstraZeneca), Dr Mark Wigglesworth (AstraZeneca), Marian Preston (AstraZeneca) | 1 comment
Increasing numbers of companies in the pharma industry are consolidating their services and outsourcing to CROs to reduce business costs. AstraZeneca’s Marian Preston, David Murray and Mark Wigglesworth discuss how this can not only drive innovation but also prove successful in identifying lead compounds, as evidenced through recent collaborations.


THE HIGH-THROUGHPUT screening (HTS) landscape has seen significant changes over the past decade. Where previously large pharmaceutical companies had multiple HTS groups located at R&D sites across the world, the current model sees many with just a single global screening centre and, in some instances, HTS groups closed altogether in favour of outsourcing large-scale screening to contract research organisations (CROs). These changes have been driven by measures to improve efficiencies, company reorganisations and portfolio refocusing and has meant that in 2019, following GlaxoSmithKline’s closure of the Harlow site in 2010 and Pfizer’s exit of the Sandwich research facility in 2011, AstraZeneca (AZ) has the only remaining pharma HTS group in the UK. Prior to this, AZ had also reduced from four global screening sites in 2010 to a single HTS centre by 2012. Yet advances in drug discovery are dependent on identification of chemical starting points for discovery projects and HTS remains the cornerstone of this operation across the pharmaceutical industry.1-3
The benefits of consolidating large-scale screening to one site are not limited to reducing the high costs of supporting multiple groups. Generation of a single facility with the capabilities to perform a vast array of assay technologies removes the need to either duplicate these at multiple sites or select specific sites for specific technologies and has the added benefit of attracting expertise in the form of dedicated screening scientists and automation engineers. This is a conclusion that has also been reported following significant investment in the National Institute of Health (NIH) in the USA, where, when reviewed in 2012,4 investments from the Molecular Libraries Program had resulted in the creation of 32 independent centres, each with distinct and limited capabilities. The recommendation was that future investments be more focused on creating fewer centres with stronger capabilities.
Large pharma generally have larger, more diverse and higher quality compound collections”
Contrary to the trend of consolidation within the pharma industry, more academic groups have established their own screening facilities to support research and drug discovery projects and, according to data from the Society for Laboratory Automation and Screening (SLAS) website, there are now more than 100 academic centres focusing on screening (small molecule and other modalities). While the number of academic screening centres has grown in recent years, each built to comprise varied technology and skill sets equivalent to pharma groups, it is rare to find a centre that has the breadth of capability available to pharmaceutical sites. Additionally, the creation of HTS screening libraries at sizes beyond 500,000 compounds is almost unheard of across these groups, with just four sites globally (two in the USA, one in India and one in Taiwan) recorded by SLAS as having greater than this number of compounds, one of which is a genomics centre (Figure 1).
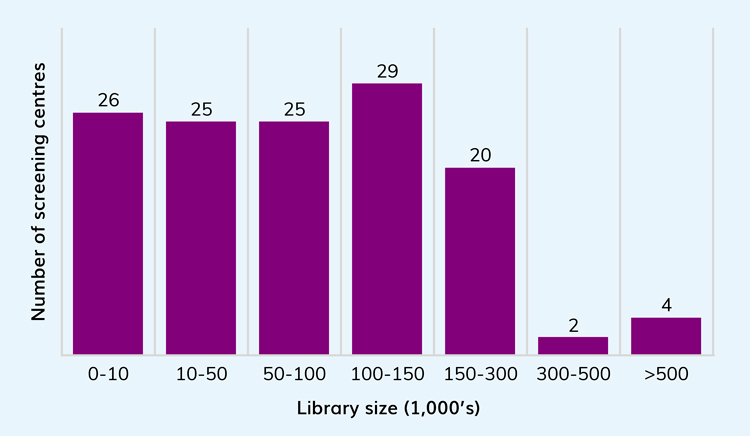

Figure 1: Library size for groups registered with SLAS. Although not a complete register of global organisations it does show a clear drop-off in numbers of organisations with access to libraries with >300K compounds.
While this may not represent all the global screening sites or capabilities, it is a strong indication that further investment in focused global centres can be made to strengthen early drug discovery and alliances between pharmaceutical companies, academic centres and CROs. This is important to expand capacity, make the best use of equipment and allow a sustainable cross fertilisation of skill sets and experience.
Open innovation
Over the last few years there has been a marked change in how large pharma companies approach drug discovery and HTS.5 Prior to this change, pharma companies tended to be inward looking, quite secretive organisations often focusing on diseases with high revenue potential and blockbuster medicines. In more recent years, it has become evident that novel drug targets, often addressing smaller patient populations and harder to treat diseases, are forming the balance of pharmaceutical portfolios. These targets are more difficult to identify and assays can be harder to develop. This has caused large pharma companies to become far more outward looking with many, if not all, of the leading companies having open innovation programmes. These programmes are designed to facilitate collaboration with academic groups and aim to drive innovation in disease target biology to identify new therapeutics. Despite the growth in academic HTS centres, they generally do not have the infrastructure to enable the running of very large and/or complex screens. Additionally, large pharma generally have larger, more diverse and higher quality compound collections and many companies are opening up access to these unique resources in an effort to discover new chemical starting points to treat the still large number of poorly treated or untreated diseases.
That said, there are some examples where these aspects are being addressed through consortia and collaborative projects. For example, the European Lead Factory6,7 has established a substantial pharmaceutical grade compound collection and, through working with an ex-pharmaceutical site to screen projects, has been able to run an equivalent process. Further to this, with improved technologies for purification and identification of natural products, investments like the National Cancer Institute’s natural product collection8 will offer new opportunities to these communities. However, access to pharmaceutical HTS centres remains a significant opportunity for the academic community. Open innovation programmes are designed to benefit both parties, with the academic partners bringing disease knowledge, novel targets and assays and the large pharma companies bringing drug discovery expertise, state-of-the-art screening centres and compound collections. Such collaborations are often set up such that the pharma company has the right of first refusal to licence any chemistry found within the joint project with the aim to co-develop a therapeutic product.
HTS open innovation in AstraZeneca
As the sole remaining pharma HTS group in the UK, AZ are uniquely positioned to be able to share their HTS and compound management infrastructure with the wider scientific community.
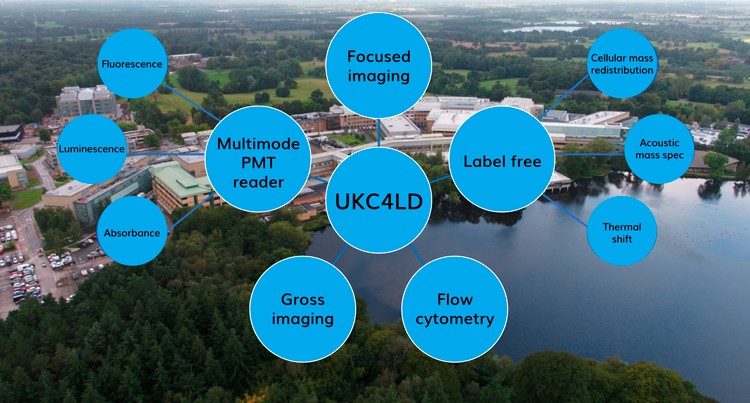

Figure 2: Assay technologies available to collaborators in the UK Centre for Lead Discovery
In recent years, the technology has become available to run more complex assays on hundreds of thousands to millions of compounds, including flow cytometry, mass spectrometry, high-content imaging and biophysical techniques. These direct and often label-free measurements result in more confidence in the chemical equity identified at an earlier stage of the drug discovery process and AZ can share access to this enhanced toolkit with their collaborators (Figure 2). The costs and space necessary to invest in the essential infrastructure and peripherals required to perform these assays at scale would be prohibitive to many academic labs, or even small CROs, hence collaborative ways to access this capability are essential to maximise the chance of success.
Several different models currently exist within AZ for running HTS collaborations, including the UK Centre for Lead Discovery (UKCLD) and the Open Innovation (OI) Portal. The UKCLD was formed in 2016 and is a collaborative venture between AZ, the Medical Research Council (MRC) and Cancer Research UK (CRUK), giving UK-based researchers and CRUK scientists an opportunity to screen their targets against the AZ compound collection. Projects are selected by and funded through the MRC and CRUK, respectively, resulting in coverage of a diverse range of disease areas and the freedom to pursue less well-validated targets.
The technology has become available to run more complex assays on hundreds of thousands to millions of compounds”
A different model exists for the OI Portal, whereby AZ can provide funding and therefore many of the projects are aligned to the internal AZ therapy areas. These applications are not limited by the geographic restrictions of funding bodies and, as such, come from groups across the world.
In addition to projects in which screening is performed in the UKCLD, there is an opportunity for researchers, with labs equipped for HTS, to utilise the AZ compound management capabilities to access a smaller subset (250K) of the AZ compound library to screen at their sites. Compound collections are one of pharma’s biggest assets and access to these is a valuable resource for academics and smaller drug discovery groups which has, until recently, been inaccessible.
For all models, whether screened at AZ or externally, dedicated AZ HTS scientists work alongside the researchers to provide guidance for all stages of the project from assay design to compound sharing. Support with data analysis allows the integrated AZ HTS and compound management IT systems to simplify a combination of assay data with sample tracking. As AZ compound structures remain blinded to the collaborators until formal sharing of the contractually agreed number of compounds, AZ chemists are assigned to each project to allow historical knowledge of the library and use of corporate analysis packages to be applied. It is this combination of the drug discovery expertise of AZ scientists with the expert disease knowledge of the academic partner that adds the highest value to the collaborative projects; that in turn helps generate high quality screening cascades to discover the best chemical equity within the compound libraries screened.
Case studies
Cancer Research UK Manchester Institute: project to identify inhibitors of isocitrate dehydrogenase 1 (IDH1) from the AZ compound collection
IDH1 mutations have been found in several cancer types, including glioma and acute myeloid leukaemia, and as such a mutated form (R132H) of IDH1 was used to screen 1.35 million compounds. The HTS utilised an Amplite-based biochemical primary screen and an orthogonal mass spectrometry assay and benefitted from the use of historical AZ screening data to aid removal of frequently active compounds. Following this, compounds were tested at 1x and 10x enzyme concentration with those showing a shift in pIC50 of more than 0.5 likely to be acting by an undesirable mechanism. In addition, a surface plasmon resonance (SPR) assay was performed to confirm direct binding to the IDH1 mutant enzyme, which was critical to the success of this project. This cascade of assays and analysis resulted in the identification of three hit series. Chemical re‑synthesis confirmed one active series, and this was built upon with the elucidation of the crystal structure showing binding in a novel allosteric site and demonstrating activity in the mutant but not the wild-type IDH1.
The collaborative project resulted in identification of a novel series of inhibitors, which has since been published.9
A·WOL consortium: a collaboration between the A·WOL consortium based at the Liverpool School of Tropical Medicine and the HTS Group at AstraZeneca
The project aim was to identify novel macrofilaricidal drugs targeting the essential bacterial symbiont (Wolbachia) of the filarial nematodes causing onchocerciasis and lymphatic filariasis.
Support with data analysis allows the integrated AZ HTS and compound management IT systems to simplify a combination of assay data with sample tracking”
An insect cell line stably infected with Wolbachia was used by the A·WOL consortium to develop a whole cell assay which, in collaboration with the team at AZ, was modified to be capable of being run at HTS scale. This complex screening process involved seven-day cell recovery, exposure of the cells to compounds for a further seven days before fixation, DNA staining (to determine toxicity) and antibody staining specific to Wolbachia. This was run as a combination of manual and automated steps utilising both Agilent BioCel and HighRes Biosolutions automation platforms. This complex cascade facilitated the screening of 1.3 million compounds from the AZ collection in just 10 weeks, generating over 20,000 hits. Chemical filtering was performed by AZ and A·WOL chemists from the University of Liverpool, to remove known frequent hitters and undesirable structures to generate a set of 6,000 compounds for further evaluation in concentration response testing. Fifty-seven clusters were identified and from these two exemplars from each cluster were tested in nematodes (in vitro Brugia malayi Mf). This cascade of assays resulted in identification of nine chemical series, which depleted the Wolbachia without toxicity observed in either the primary cell assay or subsequent whole-worm nematode screen. Having access to the expertise and automation platforms in the AZ HTS centre was vital to allow the screening of such a large number of compounds in such a long and complex assay. The output from the project is published in Nature Communications10 with the validation of the assay published in SLAS Discovery.11
MRC Laboratory of Molecular Biology
This involved a project to identify effectors of the microtubule motor dynein with the aim of identifying inhibitors, which will be valuable as tool compounds to assist in understanding the role of this motor in healthy cells and activators, which could be of therapeutic value for neurodegenerative disease.
An approach reported by Kapitein et al12 was adopted, which utilises the FRB-FKBP heterodimerisation system to recruit dynein and dynactin to labelled peroxisomes and thus drive their relocalisation to the centre of the cell. Developing this complex cell-based system into an HTS assay capable of identifying both inhibitors and activators required the specialist skills of HTS screening scientists combined with expertise in imaging data analysis and methodology. This was achieved by establishing optimal clonal cell lines and using lower throughput, higher resolution images to validate algorithms for quantifying inhibition and enhancement of peroxisome movement before transfer to the high-throughput CellInsight wide‑field imaging system. Use of a cutting edge HighRes Cart to Cart automation system allowed extended screening runs by maintaining the cell plates in optimum storage conditions while continually supplying the CellInsight imagers both in and outside of normal working hours. This procedure was key to enabling such a large screen to be run and generating high quality data.
A screen of 500,000 AZ compounds identified unique sets of compounds demonstrating either an inhibitory or activation effect on dynein-based transport. Following validation in an XC50 format, additional assays were performed to remove false positive fluorescent compounds and those compounds acting indirectly by disrupting microtubules. Further characterisation of shortlisted compounds is ongoing in the MRC Laboratory of Molecular Biology.
Cancer Research UK Cambridge Institute
Asparagine synthetase has been identified as a driver of breast cancer metastasis.13 A project from Greg Hannon’s lab at the Cambridge Institute to identify inhibitors of this enzyme from the AstraZeneca compound collection took advantage of acoustic mist ionisation mass spectrometry (AMI-MS) technology developed by AstraZeneca and Labcyte/Waters. This technology combines the label-free and direct measurement advantages of mass spectrometry with the speed and miniaturised assay volumes of acoustic dispensing.12 Utilising AMI-MS allowed differentiation between compounds acting on each side (glutamate and AMP products) of the reaction (Figure 3). This was central to enabling this project to find inhibitors with the required mechanism and would not have been possible in a more conventional biochemical assay format.
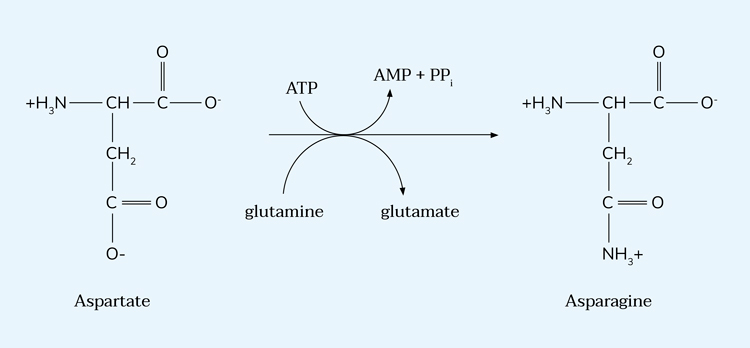

Figure 3: Schematic of the asparagine synthetase reaction
Collaborative working between teams at the Cambridge Institute, Cancer Research Technology and UKCLD enabled a 500,000-compound screen to be performed using AMI-MS. Hits were confirmed as concentration response curves and triaged using compound physicochemical properties, behaviour in previous screening campaigns, an orthogonal AMP-Glo assay and assays to identify redox cycling compounds.
An extension to the collaboration was established with the Medicines Discovery Catapult for development of a set of cell-based mass spectrometry assays. Chemical series that remained following the HTS cascade were tested in these to allow further triage, and structures have been shared for those with the best hit profiles. Work by the PI is ongoing to elucidate the MoA of these compounds.
Summary
As can be seen in the case studies above, combining disease target knowledge within the academic community with the expertise, cutting-edge technology and drug-like compound collection from AZ can lead to the discovery of new chemical starting points that can allow better understanding of the disease processes and may lead to new therapies for patients.
The future of lead discovery screening
The future of open innovation screening will, in large part, depend on the value generated from the projects and measuring this holds its own challenges – not least that this will be different for each of the invested parties. It can take many years to determine the true outcome of the projects and the measure of success will vary between individual collaborators. For some, this may be the identification and sharing of a tool compound, whereas for others it might be investment from the pharma company to develop a drug to take to market. The funding bodies are, of course, looking for a return on their investment, which in the long term will include bringing new drugs to patients, sharing scientific knowledge and advancing the understanding of disease biology through high quality publications.
With a record year for investment in biopharma in 2018 ($16.8 billion, according to EvaluatePharma), there is substantial optimism in the sector with new approaches to discover medicines and a likely growth in the demand for screening services in the next five years. It is expected that access to these capabilities through open innovation and further centralised investments will be a key platform to help discover the medicines of the future to treat more patients across more diseases than we can today.
Conclusion
Over the past decade there has been a significant increase in the number of drug discovery collaborations between academic groups/ charitable organisations and pharma. These have been formed with the aim of harnessing expertise and maximising access to cutting-edge HTS capabilities to drive forward the identification of tool and lead compounds in the battle to increase understanding of disease biology and improve lives of patients. At AZ, positive impact from early projects has been demonstrated by identification of novel chemistry and resulting publications, not only furthering the research of the collaborating academic group but also the wider scientific community.
Acknowledgment
We would like to acknowledge the skills and expertise of the large number of people involved in delivering these projects from both the academic collaborator’s laboratories and the extended Open Innovation team within AstraZeneca. Without a substantial amount of pre-work to establish these collaborations and the science behind them this publication would not have been possible.
About the authors






References
- Follmann M, Briem H, Steinmeyer A, et al. An approach towards enhancement of a screening library: The next generation library initiative (NGLI) at bayer – against all odds? Drug Discov Today. 2019;24(3):668-672.
- Wigglesworth MJ, Murray DC, Blackett CJ, Kossenjans M, Nissink JW. Increasing the delivery of next generation therapeutics from high throughput screening libraries. Curr Opin Chem Biol. 2015;26:104-110.
- Brown DG, Bostrom J. Where do recent small molecule clinical development candidates come from? J Med Chem. 2018;61(21):9442-9468.
- Wadman M. National prescription for drug development. Nat Biotechnol. 2012;30(4):309-312.
- Rees S, Gribbon P, Birmingham K, Janzen WP, Pairaudeau G. Towards a hit for every target. Nat Rev Drug Discov. 2016;15(1):1-2.
- Mullard A. European lead factory opens for business. Nat Rev Drug Discov. 2013;12(3):173-175.
- Besnard J, Jones PS, Hopkins AL, Pannifer AD. The joint european compound library: Boosting precompetitive research. Drug Discov Today. 2015;20(2):181-186.
- Thornburg CC, Britt JR, Evans JR, et al. NCI program for natural product discovery: A publicly-accessible library of natural product fractions for high-throughput screening. ACS Chem Biol. 2018;13(9):2484-2497.
- Jones S, Ahmet J, Ayton K, et al. Discovery and optimization of allosteric inhibitors of mutant isocitrate dehydrogenase 1 (R132H IDH1) displaying activity in human acute myeloid leukemia cells. J Med Chem. 2016;59(24):11120-11137.
- Clare RH, Bardelle C, Harper P, et al. Industrial scale high-throughput screening delivers multiple fast acting macrofilaricides. Nat Commun. 2019;10(1):11-018-07826-2.
- Clare RH, Clark R, Bardelle C, et al. Development of a high-throughput cytometric screen to identify anti- wolbachia compounds: The power of public-private partnership. SLAS Discov. 2019;24(5):537-547.
- Knott SRV, Wagenblast E, Khan S, et al. Asparagine bioavailability governs metastasis in a model of breast cancer. Nature. 2018;554(7692):378-381.
- Sinclair I, Bachman M, Addison D, et al. Acoustic mist ionization platform for direct and contactless ultrahigh-throughput mass spectrometry analysis of liquid samples. Anal Chem. 2019;91(6):3790-3794.
Related topics
Disease Research, Lead Generation, Research & Development, Screening
Related conditions
acute myeloid leukaemia, Cancer, Glioma
Related organisations
AstraZeneca (AZ), Cancer Research Technology, Cancer Research UK (CRUK), European Lead Factory, GlaxoSmithKline, Labcyte/Waters, Liverpool School of Tropical Medicine, Liverpool University, Medical Research Council (MRC), Medicines Discovery Catapult, National Cancer Institute, National Institute of Health (NIH), Pfizer, Society for Laboratory Automation and Screening (SLAS), UK Centre for Lead Discovery (UKCLD)







DEL will be a trend for new drugs development then HTS.