New cryo-EM method reveals insight into blood disorders
Posted: 15 February 2022 | Ria Kakkad (Drug Target Review) | No comments yet
A molecular cage, developed for use during cryo-EM, has provided researchers with new insights into a key cancer protein.

Researchers at Stanford University’s Schools of Medicine and Engineering and the Department of Energy’s SLAC National Accelerator Laboratory, US have developed a molecular cage to stabilise certain medium-sized proteins so they can be imaged for the first time with cryogenic electron microscopy (cryo-EM), which can reveal almost atomic-level details. The details of this study were recently published in ACS Central Science.
According to the researchers, there is a subtle gap between two technologies used to understand the structure and function of proteins – X-ray crystallography and cryo-EM – which has hindered their work to develop of therapeutics against acute myeloid leukaemia (AML) and other blood disorders.
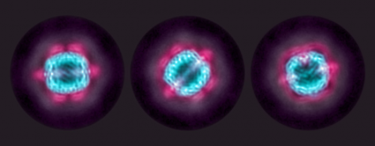

Cryo-EM reconstructions of KIX (red) sandwiched between an MBP outer shell (purple) and an apoferritin inner shell (blue). The sandwiching technique helped researchers get the best look yet of KIX, a potential target for treating acute myeloid leukaemia.
[Credit: SLAC National Accelerator Laboratory.]
A part of the issue is KIX, a segment of the CREB Binding Protein (CBP) that AML cancer cells use to transcribe genes important for growth and survival. The team say that understanding its structure better could enable them to design drugs that inhibit KIX and prevent cancer cells from replicating. However, efforts to study the protein using X-ray crystallography have not been successful; the molecule’s generous size, by crystallography standards, makes it harder to crystallise and even when it has been crystallised, the particulars of that process have made it harder to analyse the parts of KIX that drug designers would like to target.
Furthermore, KIX is too small on its own to study effectively with cryo-EM. To get good images of a protein with cryo-EM, researchers must be able to locate enough copies of the protein within an electron microscope image, then understand how they are oriented. Only by finding and lining up images of a protein can cryo-EM methods yield high-resolution structures. KIX’s small size, by cryo-EM standards, makes that a challenge. Another option, nuclear magnetic resonance, has been used to determine the structure of KIX when bound to other naturally occurring molecules, but the method requires extensive preparation and analysis, making it less than ideal for quickly determining molecule’s structures and therefore for less than ideal for studying the effects of potential KIX-inhibiting drugs.
To resolve this issue, the researchers sandwiched batches of KIX proteins between a central, ball-shaped molecule and an outer molecular cage. As this “double shell” was much larger than individual KIX molecules, it was easier to spot and orient in cryo-EM images, therefore making it easier to obtain high-resolution images of the KIX molecules themselves.
In addition to seeing KIX’s structure, the team were able to add other molecules to the mix to see if they might bind to and potentially inhibit KIX’s function. Already, the team reports, they have been able to make that bonding about 200 times stronger, which could help scientists develop drugs that are effective at lower doses.
The team’s results also suggest this method could prove useful for other proteins of in-between sizes that are hard to study with either cryo-EM or X-ray crystallography – including, perhaps, some viral proteins.
Related topics
Imaging, Microscopy, Molecular Biology, Molecular Targets, Protein, X-ray Crystallography
Related conditions
acute myeloid leukaemia (AML)
Related organisations
Stanford University








