Scientists discover extensive array of protein folds unexplored in nature
Posted: 13 July 2023 | Izzy Wood (Drug Target Review) | No comments yet
A study out of Japan reveals diversity of protein structures and how they fold in nature, uncovering a myriad of protein topologies.

A National Institutes on Natural Sciences (NINS), Japan, study has uncovered the astonishing diversity of protein structures and their folds in nature. Researchers set out to reveal the extent to which nature has explored the vast landscape of possible protein topologies. The results have unveiled an astounding array of unexplored protein folds, expanding our understanding and uncovering the depth of the protein universe. This research has been published in the journal Nature Structural and Molecular Biology.
Proteins are the building blocks of life, that fold into specific three-dimensional (3D) structures, enabling them to carry out their biological functions. The 3D structures of proteins are dictated by their amino acid sequences.
While previous experimental techniques have successfully unravelled the structures of numerous proteins, the discovery of new protein folds, defined by the arrangement and connectivity of α-helices and β-strands, has become increasingly infrequent.
This raises the question: how extensive is the protein fold space not explored by nature?
To address this question, the research team embarked on a study combining theoretical prediction for novel protein folds with experimental validation of their de novo designs.
They devised rules based on physical chemistry and protein structure data to theoretically predict possible protein folds. These rules were then employed to predict novel αβ-folds, which consist of a four to eight stranded β-sheet, not yet observed in the current Protein Data Bank (PDB). This led to the identification of a total of 12,356 novel folds. The team then attempted to computationally design proteins for the predicted novel folds from scratch to assess the foldability and fidelity of the novel folds.
“We attempted to computationally design proteins with all of the predicted folds that have a four-stranded β-sheet, including one forming a knot-like structure,” explained Shintaro Minami, a researcher at Exploratory Research Centre on Life and Living Systems (ExCELLS). “When designing proteins, we did not expect all of them, especially knot forming ones, to fold into the structures as anticipated.”
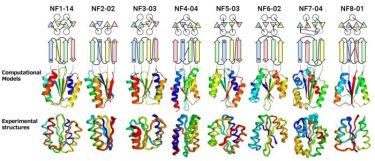

From the top, the protein name, topology diagram (circles represent α-helices, triangles and arrows represent β-strands), computationally designed structures, and experimentally determined structures. The topology of NF8-01 forms a knot. (Credit: Shintaro Minami, Naohiro Kobayashi, Toshihiko Sugiki, Toshio Nagashima, Toshimichi Fujiwara, Rie Tatsumi-Koga, George Chikenji & Nobuyasu Koga, “Exploration of novel αβ-protein folds through de novo design”, Nature Structural & Molecular Biology (2023))
The results of experimental testing were surprising (See Figure). “For all of the folds, the computationally designed protein structures closely matched the experimental structures,” said Naohiro Kobayashi, a senior research fellow at RIKEN.
These findings suggest the existence of at least approximately 10,000 unexplored foldable αβ-folds, a significant revelation considering only 400 αβ-folds have been observed in nature. This suggests that many potential folds remain uncharted in the protein folding space.
These results have given rise to several hypotheses about the structure and evolution of proteins. One hypothesis is that proteins may have not been present in biology long enough for all possible folds to have been explored.
Another hypothesis is that protein folds in nature are inherently biased due to all life on Earth having descended from a common ancestor.
“The design of proteins with these novel folds will lead to an even greater diversity of structures. This would pave the way for the de novo design of functional protein molecules, leading to breakthroughs in drug development, enzyme design, and other areas,” concluded Nobuyasu Koga, Professor at the ExCELLS, NINS.
Related topics
Amino Acids, Computational techniques, Molecular Biology, Protein, Structural Biology
Related organisations
Exploratory Research Centre on Life and Living Systems (ExCELLS), National Institutes on Natural Sciences (NINS)
Related people
Naohiro Kobayash, Nobuyasu Koga, Shintaro Minami