AI system produces binding proteins using limited target information
Posted: 20 December 2023 | Drug Target Review | No comments yet
New software can make protein molecules that bind with high affinity and specificity to many biomarkers, including human hormones.
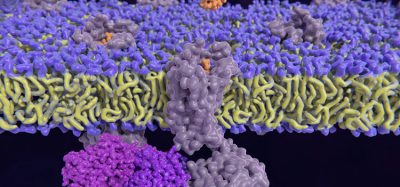
Researchers at the Institute for Protein Design at the University of Washington School of Medicine have used software to make protein molecules that bind with exceptionally high affinity and specificity to a variety of difficult biomarkers, including human hormones. They achieved the highest interaction strength ever reported between a computer-generated biomolecule and its target, which has great implications for drug development.
Dr David Baker, professor of biochemistry at UW Medicine, Howard Hughes Medical Institute investigator and senior author of the study said: “The ability to generate novel proteins with such high binding affinity and specificity opens up a world of possibilities, from new disease treatments to advanced diagnostics.”
Led by Baker Lab members Susana Vazquez-Torres, Preetham Venkatesh, and Phil Leung, the team set out to create proteins that could bind to glucagon, neuropeptide Y, parathyroid hormone, and other helical peptide targets. These molecules are essential in biological systems but are particularly challenging for drugs and diagnostic tools to recognise because they often lack stable molecular structures. Some of these medically relevant targets can be detected by antibodies but are expensive to produce and have limited shelf lives.
Venkatesh explained: “There are many diseases that are difficult to treat today simply because it is so challenging to detect certain molecules in the body. As tools for diagnosis, designed proteins may offer a more cost-effective alternative to antibodies.”
Deep-learning methods
The study introduces a new protein design approach that uses advanced deep-learning methods. The scientists used two programmes developed in the Baker lab: RFdiffusion, a generative model for creating new protein shapes, in conjunction with the sequence-design tool ProteinMPNN. This enabled them to create functional proteins more efficiently than before.
By combining these tools in new ways, the team generated binding proteins by using limited target information, such as a peptide’s amino acid sequence alone. This approach suggests a new era in biotechnology in which artificial intelligence (AI)-generated proteins can be used to detect complex molecules relevant to human health and the environment.
The team then conducted laboratory tests in collaboration with the Joseph Rogers Lab at the University of Copenhagen and the Andrew Hoofnagle Lab at UW Medicine to validate their biodesign methods. Using mass spectrometry, they found designed proteins that bind to low-concentration peptides in human serum, thereby demonstrating the potential for sensitive and accurate disease diagnostics.
The proteins retained their target binding abilities despite harsh conditions including high heat, a crucial attribute for real-world application. This method’s potential was further showed when the team integrated a high-affinity parathyroid hormone binder into a biosensor system and achieved a 21-fold increase in bioluminescence signal in samples that contained the target hormone.
This integration into a diagnostic device demonstrates the immediate practical applications of AI-generated proteins.
This study was published in Nature.
Related topics
Amino Acids, Artificial Intelligence, Biomarkers, Drug Targets, Protein
Related organisations
Copenhagen University, Howard Hughes Medical Institute, University of Washington School of Medicine