New technique developed to expand multimodal single cell analysis
Posted: 23 April 2019 | Fraser Owen (Drug Target Review) | No comments yet
A new technique, called ECCITE-seq, has been developed to allow researchers to perform high-throughput measurements of multiple modalities of information from single cells.
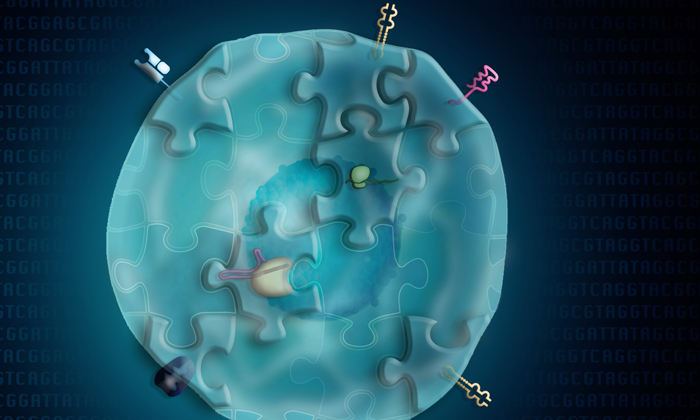

Depiction of a single cell illustrating the multimodal view of biomolecules that can be measured with ECCITE-seq
Developed by scientists at the New York Genome Centre‘s (NYGC), ECCITE-seq, (Expanded CRISPR-compatible Cellular Indexing of Transcriptomes and Epitopes by sequencing) profiles different types of biomolecules from thousands of single cells in parallel. This offers a breadth of information that can be used as readout in CRISPR-based pooled genetics screens.
“ECCITE-seq is a next-generation tool that enhances our ability to more thoroughly investigate single cells and better understand disease mechanisms,” said NYGC’s Scientific Director and Chief Executive Officer, Tom Maniatis, PhD.
This technique expands from a related tool from 2017 that the Technology Innovation Lab published (named CITE-seq) which enables the detection of proteins together with transcriptomes in single cells.
Then, in 2018, they followed up this work by using the same concept to barcode individual samples, allowing multiplexing of single cell RNA sequencing experiments in a method named Cell Hashing.
“For ECCITE-seq, we adapted our previous work on protein detection and sample multiplexing and combined them with the ability to directly detect single guide RNAs used for CRISPR screens,” added Peter Smibert, PhD, Manager of the NYGC Technology Innovation Lab and corresponding author on the new study. “The separate measurements in ECCITE-seq are modular, so researchers can pick and choose which measurements they need to make for the question they are asking.”
ECCITE-seq is built upon the Single Cell Immune Profiling solution from 10x Genomics, which enables researchers to reconstruct clonotypes of individual immune cells. The team combined protein detection with transcriptomes and clonotypes to characterise malignant populations in a sample from a patient with cutaneous T cell lymphoma.
Combining modalities enabled fine dissection of specific cell subtypes and helped reveal a transcriptomic signature of malignant cells.
“While we have demonstrated the utility of this method in cancer, it is a platform that can be applied to the study of a range of biological systems and diseases,” Dr Smibert continued. “With future developments including the addition of new modalities, we see ECCITE-seq and future methods that expand upon it serving as fundamental tools for better interrogation of individual cells.”
This research was published in Nature Methods.
Related topics
Cell Cultures, CRISPR, Genomics, RNAs, T cells
Related conditions
Cancer
Related organisations
New York Genome Centre
Related people
Peter Smibert PhD, Tom Maniatis PhD