T-box and tRNA complex structure revealed by study
Posted: 19 November 2019 | Victoria Rees (Drug Target Review) | 1 comment
A team of researchers has uncovered the structure of a T-box regulatory mechanism in bacteria, which could aid in the development of novel antibiotics.
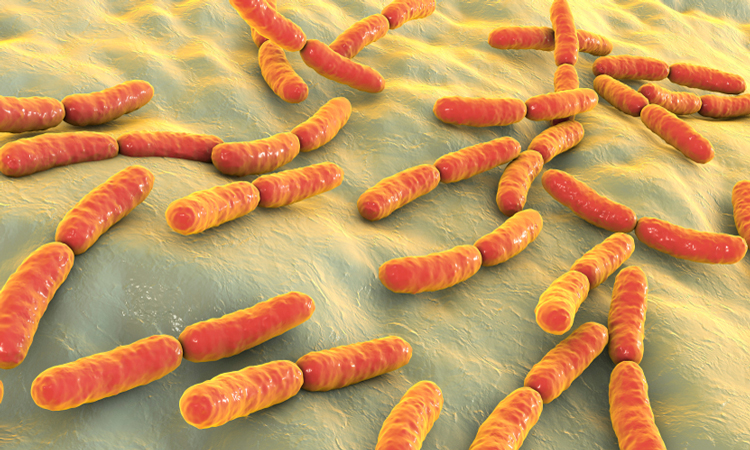

The structure of a regulatory mechanism unique to bacteria has been revealed by researchers. According to the team, their findings could present an opportunity to design new antibiotics.
The study, conducted at Cornell University, US, used X-ray crystallography to uncover the ‘T-box’ elements in Mycobacterium tuberculosis. T-boxes are the structures that recognise when a cell is deficient in a specific amino acid and allow bacteria to respond to this deficiency by initiating a process that generates more of that amino acid.
“The T-boxes are only found in bacteria and they control essential genes,” said study first author Robert Battaglia. “This makes them an attractive target because they are also essential for a lot of these bacteria to respond to starvation conditions.”
The researchers found that the T-boxes bind to an essential macromolecule called tRNA, which exists in uncharged and charged forms. Different types of tRNA are each fitted to a specific type of amino acid.
When the amount of an amino acid is low in bacteria, the corresponding tRNA will be uncharged and will bind to a T-box, which in turn discovers what type of amino acid that tRNA requires. This then facilitates a process of generating that amino acid. When the amino acid level increases, the tRNA will couple with it, thereby charging that tRNA. The amino acids are then used in all kinds of basic bacteria cell functions.
In the study, the researchers were able to describe the structure of the full T-box and tRNA complex.
“By solving our structure, we’re able to see how different parts of the T-box are positioned to allow the T-box to have this specific interaction with a tRNA,” Battaglia continued. “If we can develop some sort of drug to target these T-box elements to mess with their ability to bind with the tRNA, they could be a really good choice for an antibiotic because we don’t have [T-boxes] ourselves, so we don’t have to worry about side effects or toxicity.”
The results were published in Nature Structural & Molecular Biology.
Related topics
Antibiotics, Drug Targets, Imaging, Microbiology, Research & Development, RNAs
Related organisations
Cornell University
Related people
Robert Battaglia







Covid19 is very dangerous.
I hope scientists can solve this problem.