3D models developed by researchers to optimise protein design
Posted: 28 February 2020 | Victoria Rees (Drug Target Review) | No comments yet
A computational programme has been created by researchers to aid in the design of proteins for therapeutics, to predict interactions.
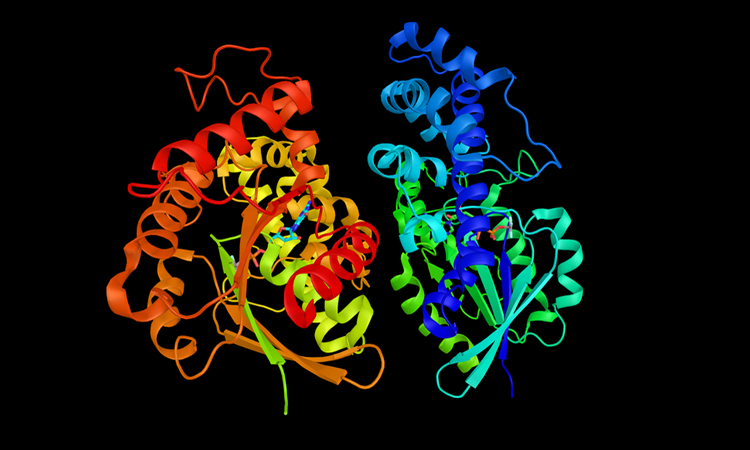

Researchers have developed a process that they say could reduce the work involved in computational protein design. The technique uses three-dimensional (3D) structural models to project how novel combinations of molecular blocks might work together to achieve a desired effect.
The team, from Dartmouth College, US, suggest that focusing on a relatively small number of protein substructures rather than the infinite number of atomic-level combinations, could ease the development of new medications and materials.
…a small number of structural patterns frequently recurred in proteins and much of the diversity in protein structure comes from how these building blocks are combined”
“When you design a building, you don’t necessarily need to understand how grains of sand interact with each other within one brick,” said Gevorg Grigoryan, an associate professor of computer science at Dartmouth and senior researcher on the study. “Because you know what a brick is and what its properties are, you can instead focus on how bricks come together to form the desired shape. That’s the same approach we are taking. We only focus on protein sub-structures that we know work.”
While many therapeutics like insulin are produced from naturally occurring proteins, the custom field has not advanced to allow widespread development of synthetic proteins. Among the barriers to developing synthetic proteins is the overwhelming number of possible amino acid combinations. Sorting through combinations to find one that would be helpful in any given scenario is a time-intensive and resource-heavy process.
Researchers developing new drugs currently focus on how specific atoms interact. This approach requires labs to build large libraries of variants to find one that will complete the specified task. While this can produce useful results, researchers have found it challenging to build atomic models that have high levels of accuracy.
“The number of sequences is virtually infinite. This really complicates the process of finding a correct combination to fill a specific therapeutic need,” said Jianfu Zhou, a PhD student at Dartmouth who co-authored the research paper.
To develop an optimised approach to design, the research team scanned a database of the 3D models of 150,000 known proteins. They discovered that a small number of structural patterns frequently recurred in proteins and that much of the diversity in protein structure comes from how these building blocks are combined.
This discovery led the team to hypothesise that rather than modelling proteins as complex networks of interacting atoms, they can instead represent them much more simply as groupings of a limited set of structural building blocks.
With the new method, novel protein structures can be more easily judged against established patterns, according to the researchers. The approach allows scientists to easily experiment with more creative designs by enabling them to reference against a library of known structures.
“This technique takes the challenge away from getting the physics absolutely right at the atomic scale, potentially making computational protein design a much more robust process. Our findings should throw the doors for machine learning in protein design wide open,” said Grigoryan.
The new process focuses on the larger blocks of atoms that occur in proteins, known as tertiary motifs, to design functioning proteins. These are recurring structural arrangements – similar to an archway or column in a building – that can be applied to designing novel proteins without regard to their atomic-level composition.
Since the structures only come together in certain ways, researchers would no longer need to do the atomic-level guesswork. Instead, they would only need to focus on the blocks that fit together, ignoring those structures that would not form a functioning protein.
According to the research paper, the results “strongly argue that the Protein Data Bank is now sufficiently large to enable proteins to be designed by using only examples of structural motifs from unrelated proteins.”
By applying the new technique, the research team hopes to cut out the redundancy of rediscovering physical principles in protein structure by simply relying on those principles in the first place.
The study was published in Proceedings of the National Academy of Sciences.
Related topics
Drug Discovery, Drug Targets, Informatics, Protein, Protein Expression, Proteomics
Related organisations
Dartmouth College
Related people
Gevorg Grigoryan, Jianfu Zhou