Spike protein analysis explains faster spread of SARS-CoV-2 variants
Posted: 18 March 2021 | Victoria Rees (Drug Target Review) | No comments yet
Using cryo-electron microscopy, researchers have imaged how the SARS-CoV-2 Spike protein changes with the D614G mutation to enable faster spread of infection.
A group of researchers has analysed how the SARS-CoV-2 Spike (S) proteins changes with the D614G mutation and showed why these variants are able to spread more quickly. The D614G mutation appears on the UK, South Africa and Brazil coronavirus variants.
Led by researchers at Boston Children’s Hospital, US, the team imaged the S protein with cryo-electron microscopy (cryo-EM), which has resolution down to the atomic level. They found that the D614G mutation – substitution of in a single amino acid “letter” in the genetic code for the S protein – makes the S protein more stable when compared with the original SARS-CoV-2 virus. As a result, more functional S proteins are available to bind to angiotensin-converting enzyme 2 (ACE2) receptors, making the virus more infectious.
According to the researchers, in the original coronavirus, the S proteins would bind to the ACE2 receptor and then dramatically change shape, folding in on themselves. This enables the virus to fuse its membrane with host own cells’ membranes and get inside. However, the proteins would sometimes prematurely change shape and fall apart before the virus could bind to cells. While this slowed the virus down, the shape change also made it harder for the immune system to contain the virus.
“Because the original S protein would dissociate, it was not good enough to induce a strong neutralising antibody response,” said Dr Bing Chen, lead researcher of the study.
When the team imaged the mutant S protein, they found that the D614G mutation stabilises the protein by blocking the premature shape change. The mutation also makes the S proteins bind more weakly to the ACE2 receptor, but the fact that the proteins are less apt to fall apart prematurely renders the virus overall more infectious.
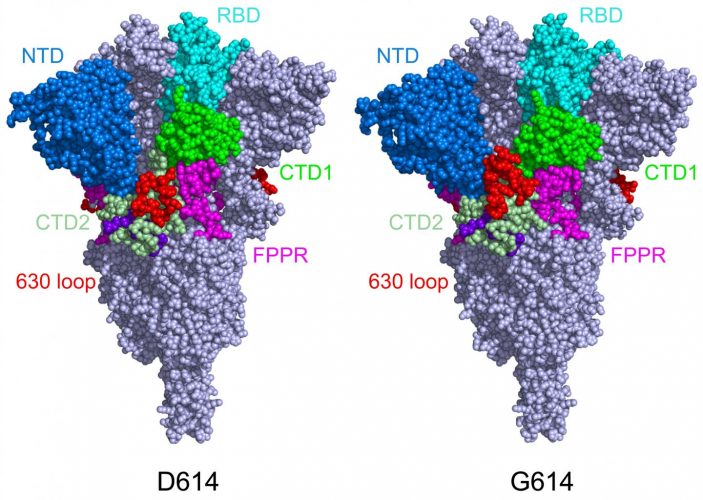
This model shows the structure of the S protein in its closed configuration, in its original D614 form (left) and its mutant form (G614). In the mutant S protein, the 630 loop (in red) stabilises the S, preventing it from flipping open prematurely and rendering SARS-CoV-2 more infectious [credit: Dr Bing Chen, Boston Children’s Hospital].
“Say the original virus has 100 S proteins,” Chen said. “Because of the shape instability, you may have just 50 percent of them functional. In the G614 variants, you may have 90 percent that are functional so even though they do not bind as well, the chances are greater that you will have infection.”
Chen proposes that redesigned vaccines incorporate the code for this mutant S protein. The more stable S protein shape should make any vaccine based on the protein more likely to elicit protective neutralising antibodies.
The findings are published in Science.
Related topics
Disease Research, Immunology, Microscopy, Vaccine
Related conditions
Covid-19
Related organisations
Boston Children's Hospital
Related people
Dr Bing Chen