Targeting gene regulation may hold key to future Alzheimer’s therapies
Posted: 10 September 2025 | Drug Target Review | No comments yet
Researchers have discovered that Alzheimer’s disease is driven by a deeper loss of gene regulation in brain cells – offering potential new targets for future therapies.

Alzheimer’s disease is known for causing devastating memory deterioration. While many new drugs aim to target features of the disease like amyloid plaques, researchers at MIT have found that targeting the regulation of gene expression in brain cells could be a more effective method.
A new study, published in Cell, provides a broad view of Alzheimer’s progression, showing how the ability – or inability – of brain cells to regulate their genes determines the loss or preservation of function and cognition.
A first-of-its-kind atlas
The researchers built a multimodal atlas spanning 3.5 million cells from six brain regions. By profiling 384 post-mortem brain samples from 111 donors, they analysed both the transcriptome (which genes are expressed into RNA) and the epigenome (chromosomal modifications that regulate DNA accessibility).
Reduce preclinical failures with smarter off-target profiling
24 September 2025 | 15:00PM BST | FREE Webinar
Join this webinar to hear from Dr Emilie Desfosses as she shares insights into how in vitro and in silico methods can support more informed, human-relevant safety decisions -especially as ethical and regulatory changes continue to reshape preclinical research.
What you’ll learn:
- Approaches for prioritizing follow-up studies and refining risk mitigation strategies
- How to interpret hit profiles from binding and functional assays
- Strategies for identifying organ systems at risk based on target activity modulation
- How to use visualization tools to assess safety margins and compare compound profiles
Register Now – It’s Free!
“This is the first large-scale single-cell multi-region gene-regulatory atlas of AD, systematically dissecting the dynamics of epigenomic and transcriptomic programs across disease progression and resilience,” said senior author Manolis Kellis, professor in MIT’s Computer Science and Artificial Intelligence Lab and head of the Computational Biology Group.
The atlas discovered two key epigenomic trends in Alzheimer’s:
- Breakdown of nuclear compartments: they normally separate active and inactive genomic regions.
- Loss of epigenomic information: meaning cells lose their unique regulatory patterns and identity.
When regulation fails, cognition declines
The study linked epigenomic breakdown with cognitive decline. Cells that maintained stable gene regulation could suppress disease-associated genes – preserving cognition. Similarly, where regulation eroded, harmful genes became active and cognition deteriorated.
Cells that maintained stable gene regulation could suppress disease-associated genes – preserving cognition.
“To understand the circuitry, the logic responsible for gene expression changes in Alzheimer’s disease, we needed to understand the regulation and upstream control of all the changes that are happening and that’s where the epigenome comes in,”Kellis explained.
Co-corresponding author Li-Huei Tsai, Picower professor and director of The Picower Institute for Learning and Memory, added: “The key to developing new and more effective treatments for Alzheimer’s disease depends on deepening our understanding of the mechanisms that contribute to the breakdowns of cellular and network function in the brain. This new data advances our understanding of how epigenomic factors drive disease.”
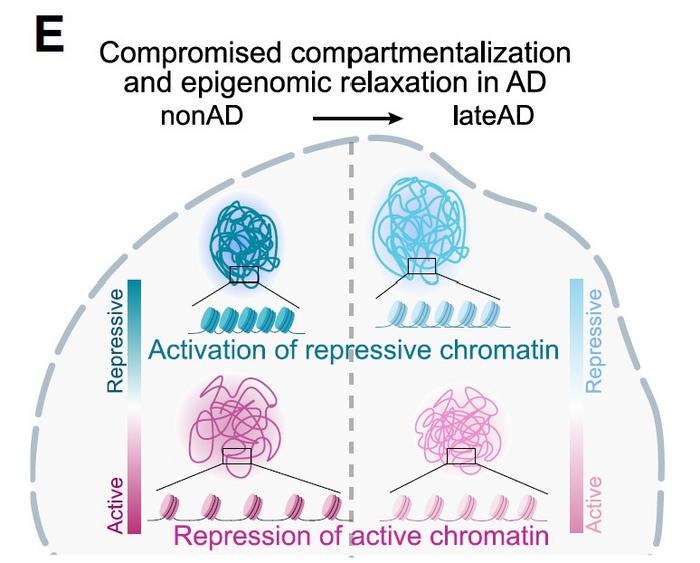

A figure from the paper illustates a key finding of “compromised compartmentalization”: chromatin that was locking genes down in health became more open in Alzheimer’s, while chromatin that was open became more locked down. Credit: Kellis & Tsai Labs/MIT
Compromised compartments and eroded information
The donor samples came from participants in long-running studies of ageing and cognition. Fifty-seven donors showed no Alzheimer’s pathology, thirty-three were at an early stage and twenty-one at a late stage.
By combining single-cell RNA sequencing with ATACseq (which maps chromatin accessibility), the team identified over 1 million gene-regulatory control regions.
By combining single-cell RNA sequencing with ATACseq (which maps chromatin accessibility), the team identified over 1 million gene-regulatory control regions. Comparing healthy and diseased brains showed how epigenomic erosion correlated with pathology and symptoms.
“For Alzheimer’s patients, repressive compartments opened up and gene expression levels increased, which was associated with decreased cognitive function,” said first author Zunpeng Liu.
Importantly, cells that managed to maintain normal compartmentalisation preserved cognitive resilience.
Vulnerable brain regions and cell types
The erosion of epigenomic information was most severe in the entorhinal cortex and hippocampus – regions that are hit earliest by Alzheimer’s. Specific vulnerable cell types included:
- Microglia: immune-supporting cells.
- Oligodendrocytes: which insulate neurons.
- Excitatory neurons: especially those expressing the RELN gene.
For example, RELN-expressing neurons suffered early information loss in most patients, but in cognitively resilient individuals, they maintained their epigenomic integrity.
Risk genes and ‘chromatin guardians’
The study also underlined how risk genes interact with epigenomic erosion. For example, the APOE4 variant, the biggest known genetic risk factor for Alzheimer’s, destabilised microglia. Initially, microglia increased their regulatory activity in response to pathology, but as the disease progressed, they collapsed – especially in individuals with two APOE4 copies.
The message is clear: Alzheimer’s is not only about plaques and tangles, but about the erosion of nuclear order itself
The researchers also identified so-called ‘chromatin guardians’ – genes that help preserve nuclear order. When these guardians faltered, cells shifted from expressing resilience-promoting genes to activating pathways linked to inflammation and stress.
“The message is clear: Alzheimer’s is not only about plaques and tangles, but about the erosion of nuclear order itself,” Kellis said. “Cognitive decline emerges when chromatin guardians lose ground to the forces of erosion, switching from resilience to vulnerability at the most fundamental level of genome regulation. When our brain cells lose their epigenomic memory marks and epigenomic information at the lowest level deep inside our neurons and microglia, it seems that Alzheimer’s patients also lose their memory and cognition at the highest level.”
A blueprint for future therapies
By mapping how epigenomic erosion drives Alzheimer’s progression, the study offers a framework for future treatments – whether targeting broad epigenomic stability or specific vulnerabilities in neurons and glial cells. This study could move the future focus of Alzheimer’s research from plaques and tangles to the deepest levels of genome regulation.
Related topics
Central Nervous System (CNS), Disease Research, Epigenetics, Neurosciences, Next-Generation Sequencing (NGS), Sequencing, Translational Science
Related conditions
Alzheimer's disease (AD)
Related organisations
MIT