Five of the most recent microscopy developments
Posted: 23 July 2020 | Victoria Rees (Drug Target Review) | No comments yet
Drug Target Review highlights five of the latest imaging advancements in the field of microscopy.


Microscopy is an imaging technique used by a range of scientists, including for the study of organic materials. The method can therefore have wide-ranging impacts for biomedical research, revealing biomarkers and drug targets.
Here, Drug Target Review lists five of the latest studies that have advanced the technique to improve imaging even further.
The structure of amyloid proteins
Researchers from Washington University in St Louis, US, developed an optical microscopy technique that can reveal nanoscale details about the structures of misfolded amyloid proteins in the brain – a hallmark of Alzheimer’s and Parkinson’s disease.
The team created several single-molecule super-resolution microscopy methods that measure the orientation and location of fluorescent molecules attached to single proteins. According to the scientists, the orientation information is obtained by measuring not only the location of fluorescence in the sample but also characteristics of that light, such as polarisation.
In their article, the researchers describe a performance metric they designed to characterise how sensitively various microscopes can measure orientations of fluorescent molecules. Using the new performance indicator, they found that a microscope which splits fluorescence light into two polarisation channels (x and y) provides superior and practical orientation measurements.
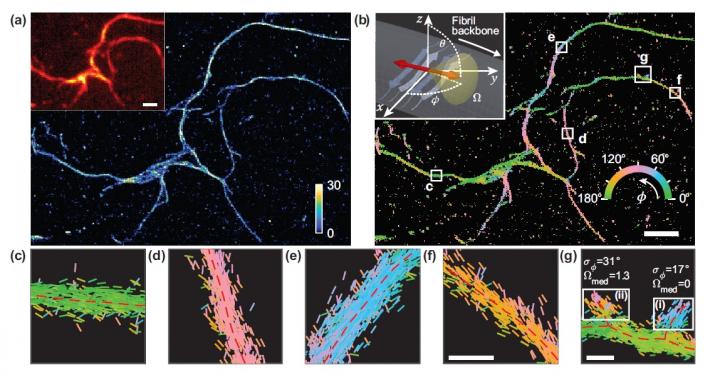

Researchers developed a microscopy technique that measures the location and orientation of single molecules and used it to study the structural details of amyloid protein aggregates. (a) Single-molecule localisation microscopy image of a network of amyloid aggregates. (b) Image showing Nile red binding orientations to amyloid surfaces, colour-coded according to the average orientation measured within each bin. (c-g) Individual orientation measurements localised along fibril backbones within the white boxes in (b). The lines are oriented and colour-coded according to the direction of the estimated angle. Horizontal white scale bars are length markers, 1 micron in (a), (b) and 100 nm in (f), (g) [credit: Tianben Ding, Tingting Wu and Matthew D. Lew, Washington University in St Louis].
The scientists say that the metric they developed calculates the performance of a particular microscope design 1,000 times faster than before.
As there is no artificial link between the fluorescent probes and amyloid surfaces, the probes’ binding orientation to the amyloid surfaces conveys information about how the amyloid protein itself is organised. The researchers quantified how the orientations of fluorescent molecules varied each time one attached to an amyloid protein. Therefore, the differences in these binding behaviours can be attributed to structure differences between amyloid aggregates.
As this method provides single-molecule information, the researchers say they could observe nanoscale differences between amyloid structures without averaging out details of local features. They now plan to conduct long-term studies on the correlations between how amyloid proteins are organised and how quickly they grow or spontaneously dissolve.
The study was published in Optica.
Microscopy for nanoscale objects
New University of Illinois, US, research has progressed optical microscopy, giving the field a new tool to solve challenging problems across many fields.
According to the team, their study will allow researchers to directly visualise unlabelled objects that have deep sub-wavelength separations. It means scientists can see nanoscale structures without performing any image post-processing.
The work began in 2018, when the researchers were conducting a theoretical study on wafer defect inspection and needed to build a simulation tool to model how light propagates through a microscope system. When they saw the simulation result for one of the configurations, they began to develop a closed form analytic expression that could explained the findings and test their hypotheses.
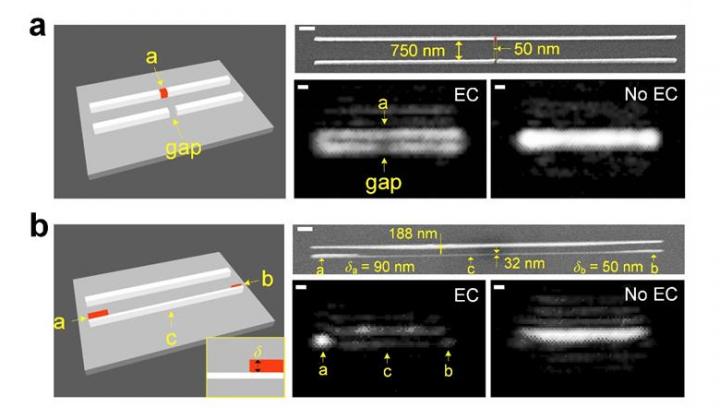

Experimental visualisation of individual nanowires and their and fabrication imperfections. The new and conventional optical microscope methods are labelled (EC) and (No EC), respectively [credit: Lynford Goddard, Grainger Engineering].
The team eventually realised the necessary experimental conditions and directly visualised their first set of deep sub-wavelength objects. They explain that their experiment utilised anti-symmetric excitation and non-resonance amplification to boost the signal-to-noise ratio of the nanoscale objects.
The team demonstrated the technique can sense both free-form and fixed-form nanoscale objects across a wide field of view (726-μm × 582-μm) using a low numerical aperture objective (0.4 NA). Some of the nanowires on the test sample had fabrication imperfections. The researchers say this allowed them to demonstrate the visualisation of sub-20 nm defects in a semiconductor chip.
In the future, the researchers say the technique could be applied for the visualisable sensing of biological objects by choosing nanowires with optimised geometry and proper refractive index and patterning functional groups around nanowires. Once target analytes are trapped, they act as objects that may be directly visualised from the optical images.
The work was published in Nature Communications.
Fibre laser microscopy
Another team from the University of Hong Kong (HKU), in collaboration with Bielefeld University in Germany, developed a compact fibre laser microscope that they say generates far less noise than customary designs and is also compact and stable.
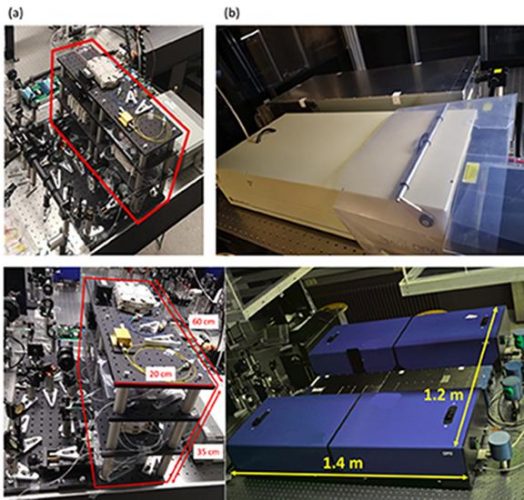

(a) Compact fibre laser sources for CRS imaging (in red box). (b) The commercial laser sources for CRS imaging [credit: The University of Hong Kong].
Furthermore, the newly developed technique does not require fluorescent markers to obtain images of cell molecules, eliminating the potential of label-induced toxicity. Instead, the method makes use of a Raman Imaging System.
According to the researchers, they used fibre laser as the light source for the microscope to replace the traditional solid-state laser. They highlight that with fibre lasers, light is amplified and transmitted through glass fibres, allowing the instrument design to become light and compact. Also, the light source uses harmless infrared visible light and will not affect the human body.
They explain that to obtain molecule-specific imaging with their microscope, the team used two synchronised optical resonators (laser cavities), both with short picosecond pulses – one picosecond being one thousand billionth of a second.
The researchers say their prototype of the microscope will now serve as the basis from which to build portable devices.
The study was published in Light: Science and Applications.
Nanoscale chemical imaging
Researchers at the Beckman Institute for Advanced Science and Technology at the University of Illinois at Urbana–Champaign, US, say they have developed a new method to improve the detection ability of nanoscale chemical imaging using atomic force microscopy (AFM). These improvements reduce the noise that is associated with the microscope, increasing the precision and range of samples that can be studied.
AFM can scan the surfaces of materials but cannot easily identify molecular composition. Researchers have previously developed a combination of AFM and infrared spectroscopy called AFM-IR. The AFM-IR microscope uses a cantilever, a beam connected to a support at one end and a sharp tip at the other, to measure subtle movements of the sample introduced by shining an IR laser. The absorption of light by the sample causes it to expand and deflect the cantilever, generating an IR signal. However, this technique produced a source of noise that limits the quality of the data.
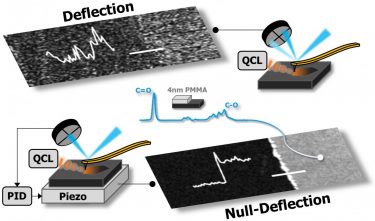

Chemical signal produced by a 4nm thick polymer film collected using previous deflection AFM-IR detection, top, compared to the new null-deflection approach [credit: Beckman Institute for Advanced Science and Technology].
The researchers created a theoretical model to understand how the instrument works and therefore identify the sources of noise. Additionally, they developed a new way to detect the IR signal with improved precision.
The researchers say that cantilever deflection is susceptible to noise which becomes worse as the deflection increases. Instead of detecting cantilever deflection, the researchers used a piezo component as a stage to maintain zero deflection. By applying a voltage to the piezo material, they were able to maintain small deflection with low noise while recording the same chemical information now encoded in the piezo voltage.
Rather than moving the cantilever, the researchers used the movement of the piezo crystal to record the IR signal.
According to the team, now that they have been able to improve the technique’s sensitivity, they can image a much smaller volume of samples, like cell membranes. They say that this method could be used to look at complex mixtures that are present in small volumes, such as a single lipid bilayer.
The results can be found in Nature Communications.
Computational microscopes
Scientists at the US National Institutes of Health (NIH), along with collaborators at the University of Chicago, US, and Zhejiang University, China, have developed new image processing techniques for microscopes that can reduce post-processing time up to several thousand-fold.
The researchers modified the deconvolution algorithm used by many so the microscope would run faster. This approach was originally proposed for other areas of medical imaging such as computed tomography (CT); however, the team say this is the first time it has successfully been adapted for use with fluorescence microscopy.
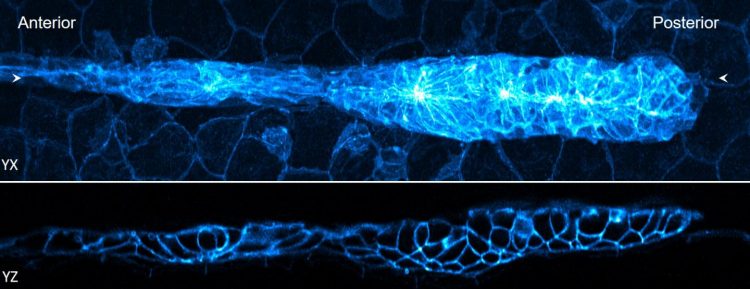

Two views of a 32-hour old zebrafish embryo. The blue fluorescence shows the edges of the cells [credit: National Institute of Biomedical Imaging and Bioengineering].
The team also reduced the time needed to position and stitch together multiple views of a sample. A key part of this advance relied on a process called parallelisation, an approach sometimes used in supercomputing where instead of processing each individual function one after another, the task is broken up into smaller tasks that can be analysed concurrently.
Finally, they showed that they could further reduce the time it takes to process the data by using a neural network, a kind of artificial intelligence (AI). The researchers trained the neural network to produce cleaner and higher resolution images much more quickly than would be possible otherwise.
The findings were published in Nature Biotechnology.
Related topics
Imaging, Informatics, Label-Free, Microscopy, Research & Development, Technology
Related conditions
Alzheimer's disease (AD), Parkinson's disease