Novel 3D imaging technology could improve fluorescence microscopy
Posted: 15 April 2020 | Victoria Rees (Drug Target Review) | No comments yet
A new technique called Coded Light-sheet Array Microscopy (CLAM) has been developed by researchers to improve 3D imaging of living specimens.


Coded Light-sheet Array Microscopy (CLAM) [credit: The University of Hong Kong].
Scientists have developed a new optical imaging technology called Coded Light-sheet Array Microscopy (CLAM) which can perform three-dimensional (3D) imaging at high speed. According to the researchers, from the University of Hong Kong (HKU), the technique is also power efficient and gentle to preserve living specimens during scanning at a level that is not achieved by existing technologies.
“CLAM allows 3D fluorescence imaging at high frame rate comparable to state-of-the-art technology (~10’s volumes per second). More importantly, it is much more power efficient, being over 1,000 times gentler than the standard 3D microscopes widely used in scientific laboratories, which greatly reduces the damage done to living specimens during scanning,” explained lead researcher Dr Kevin Tsia, Associate Professor of the Department of Electrical and Electronic Engineering at HKU.
Existing 3D biological microscopy platforms are slow because the entire volume of the specimen has to be sequentially scanned and imaged point-by-point, line-by-line or plane-by-plane. In these platforms, a single 3D snapshot requires repeated illumination on the specimen. The specimens are often illuminated by light thousands to a million times more intense than the sunlight, which is likely to damage the specimen itself.
Furthermore, these platforms often quickly exhaust the limited fluorescence ‘budget’ – a fundamental constraint that fluorescent light can only be generated upon illumination for a limited period before it permanently fades out in a process called ‘photo-bleaching’, which sets a limit to how many image acquisitions can be performed on a sample.
CLAM transforms a single laser beam into a high-density array of ‘light-sheets’ with the use of a pair of parallel mirrors, to spread over a large area of the specimen as fluorescence excitation.
“The image within the entire 3D volume is captured simultaneously, without the need to scan the specimen point-by-point or line-by-line or plane-by-plane as required by other techniques. Such 3D parallelisation in CLAM leads to a very gentle and efficient 3D fluorescence imaging without sacrificing sensitivity and speed,” said Dr Yuxuan Ren, a postdoctoral researcher of the work.
To preserve the image resolution and quality in CLAM, the team turned to Code Division Multiplexing (CDM), an image encoding technique which is widely used in telecommunication for sending multiple signals simultaneously.
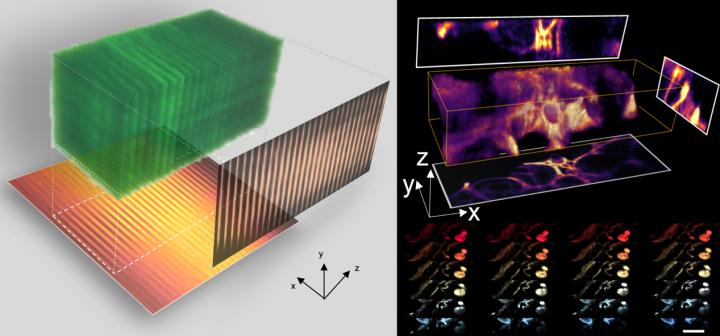

3D imaging at high speed with CLAM [credit: The University of Hong Kong].
“This encoding technique allows us to use a two-dimensional (2D) image sensor to capture and digitally reconstruct all image stacks in 3D simultaneously. CDM has never been used in 3D imaging before. We adopted the technology, which became a success,” said Dr Queenie Lai, a postdoctoral researcher who developed the system.
As a proof-of-concept demonstration, the team applied CLAM to capture 3D videos of fast microparticle flow in a microfluidic chip at a volume rate of over 10 volumes per second comparable to state-of-the-art technology.
The team combined CLAM with HKU’s Faculty of Medicine’s newly developed tissue clearing technology to perform 3D visualisation of mouse glomeruli and intestine blood vasculature in high frame-rate.
“We anticipate that this combined technique can be extended to large-scale 3D histopathological investigation of archival biological samples, like mapping the cellular organisation in the brain for neuroscience research,” Dr Tsia said.
“Since CLAM imaging is significantly gentler than all other methods, it uniquely favours long term and continuous ‘surveillance’ of biological specimen in their living form. This could potentially impact our fundamental understanding in many aspects of cell biology,” Dr Tsia added.
The study has been published in Light: Science & Applications.
Related topics
Analytical Techniques, Disease Research, Imaging, Microscopy, Research & Development
Related organisations
University of Hong Kong (HKU)
Related people
Dr Kevin Tsia, Dr Queenie Lai, Dr Yuxuan Ren