Acquiring structural data on membrane proteins – a spotlight on GPCRs
Posted: 17 November 2020 | Victoria Rees (Drug Target Review) | No comments yet
Lan Zhu from Arizona State University explains how cryo-EM methods can be used to obtain structural information on membrane proteins such as GPCRs.
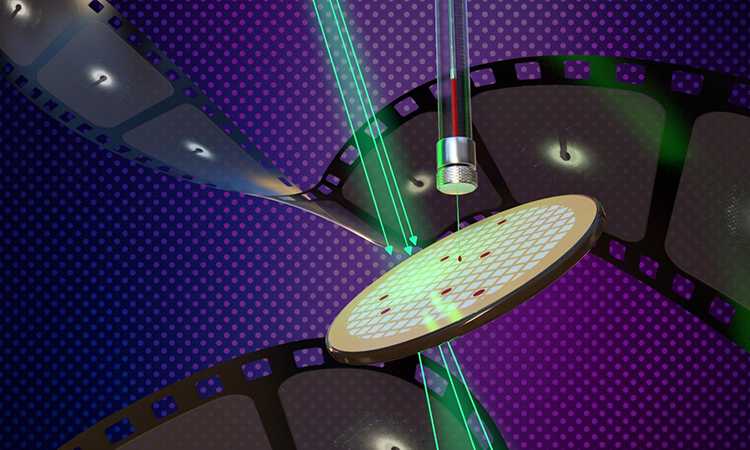

New methods of determining the structure of membrane proteins using lipidic cubic phase (LCP) microcrystals and microcrystal electron diffraction (MicroED) are described in the new study appearing on the cover of the Cell Press journal, Structure [credit: Jason Drees for the Biodesign Institute at Arizona State University].
Between 80,000 and 400,000 proteins circulate in the body’s cells, tissues and organs, carrying out a broad range of duties essential for life. However, when these proteins go awry, they can be responsible for many serious diseases.
Now, researchers at the Biodesign Center for Applied Structural Discovery and Arizona State University’s (ASU) School of Molecular Sciences have investigated a critically important class of proteins called G-protein-coupled receptors (GPCRs), which are found on the outer membranes of cells. For their study, published in Structure, the team designed a new imaging approach for acquiring structural data of membrane proteins in high detail.
Assistant Research Scientist Lan Zhu from ASU spoke with Drug Target Review about this study and why improving imaging techniques can aid our understanding of membrane proteins.
Why study membrane proteins?
Zhu said that membrane proteins comprise about 30 percent of all the proteins in eukaryotic cells and are “critical to the cell environment” due to their control over what enters cells. Other than their important biological values, the position of membrane proteins renders them ideal targets to transmit oral medications into lesions in human bodies, Zhu highlighted.

The traditional method
Emphasising the importance of obtaining structural information on these proteins, Zhu quoted the words of Dr Martin Caffrey: “If we say a picture is worth a thousand words, then a structure is actually worth a million.”
She explained that the method most commonly used to study proteins is X-ray crystallography. This involves striking crystallising protein samples with an X-ray beam, causing diffraction patterns that appear on a screen. Assembling thousands of these diffraction snapshots allows for a high-resolution three-dimensional (3D) structural image to be assembled with the aid of computers.
…the crystallisation of many membrane proteins remains extremely difficult and imprecise”
However, the researchers emphasise that many membrane proteins, including GPCRs, experience challenges to form large, well-ordered crystals appropriate for X-ray crystallography. Furthermore, such proteins are delicate and easily damaged by X-radiation. Getting around the problem has required the use of special devices known as X-ray free electron lasers (XFELS), which can deliver a burst of X-ray light lasting only a few femtoseconds (equal to one quadrillionth of a second). Therefore, the technique of serial femtosecond X-ray crystallography has allowed researchers to obtain refraction images before the crystalised sample is destroyed.
Despite this, the researchers highlight that the crystallisation of many membrane proteins remains extremely difficult and imprecise and as there are only a handful of XFEL machines in the world, other imaging techniques are required.
An improved technique
So, the researchers turned to cryogenic electron microscopy (cryo-EM) methods, microcrystal electron diffraction (MicroED) and lipidic cubic phase (LCP) crystallisation. Zhu explained that this technique involved flash-freezing protein crystals in a thin veneer of ice, then subjecting the samples to a beam of electrons. As in the case of X-ray crystallography, the method used diffraction patterns to assemble final detailed structures; however, this time the diffraction came from electrons rather than X-rays.

Zhu highlighted that LCP crystallisation has demonstrated its many benefits for membrane protein structure determination in the past few decades, while MicroED can collect data from crystals too small and irregular to be used for conventional synchrotron X-ray crystallography. In their study, the researchers used MicroED after LCP crystallisation to produce high-resolution diffraction images of two important model proteins: Proteinase K, and the human A2A adenosine receptor whose functions include modulation of neurotransmitters in the brain, cardiac vasodilation and T-cell immune response.
First, the researchers embedded the proteins in LCP, which mimics the native environment such proteins naturally occur in by providing a lipid environment very close to that of the native membrane protein. However, Zhu explained that with the high viscosity of the LCP matrix, the small crystals are not well suited to standard MicroED sample preparation protocols. To overcome this challenge, the researchers lowered the viscosity by mixing the sample with certain chemicals to convert the phase into a less viscous phase while the second was to utilise the lipase and hydrolyse the LCP matrix to convert it to a liquid phase.
The LCP samples were then subjected to electron microscopy, using the MicroED method, which permitted the imaging of the extremely thin, sub-micron-sized crystals. The team then continually rotated the LCP crystals under the electron microscope to allow multiple diffraction patterns to be acquired from a single crystal with an extremely low, damage-free electron dose. “With the new methodology, we do not have to optimise the crystal size,” Zhu said.
…the position of membrane proteins renders them ideal targets to transmit oral medications”
The team conclude that the ability to examine proteins that can only form micro- or nanocrystals opens the door to the structural determination of many vitally important membrane proteins that have eluded conventional means of investigation, particularly GPCRs. The extensive application of LCP-MicroED offers promise for improved approaches for structural determination of challenging protein targets. Emphasising the importance of the new LCP method, Zhu remarked that as only a few crystals are needed, it makes the process much more accessible to researchers and that overall, these blueprints can be used to facilitate new therapeutic drug design from more precise insights.
Related topics
Crystallography, Imaging, Microscopy, Protein, Proteomics, X-ray Crystallography
Related organisations
Arizona State University (ASU)
Related people
Dr Martin Caffrey, Lan Zhu