Droplet Digital PCR detects and quantifies cancer mutation
Posted: 31 October 2018 | Drug Target Review | No comments yet
Two ddPCR-based assays were developed to detect the two mutations in the promoter region of the TERT gene, which could be used as biomarkers…
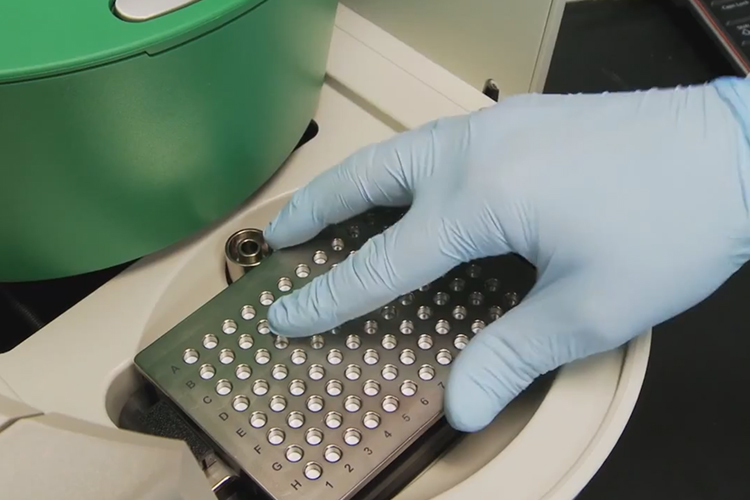

Researchers have demonstrated that Droplet Digital PCR (ddPCR) technology can be used to detect mutations in a region of DNA that is linked to many cancer types.
Two mutations in the promoter region of the TERT gene are prevalent in many cancers, with the mutations ranging from 10 to 70 percent frequency in melanoma, glioblastoma, and thyroid and liver cancers, and could also be used as a biomarker.
“We wanted to develop ddPCR assays for these TERT promoter mutations since they are very frequently mutated in melanomas, including the one third of melanoma patients who are lacking BRAF and NRAS mutations,” said Dr David Polsky, Professor of Dermatologic Oncology at NYU Langone and a senior investigator on this study. “We hope that multiple groups can use these assays in various cancer types and that detecting and quantifying these TERT promoter mutations will be useful in the evaluation and management of cancer patients.”
Automation now plays a central role in discovery. From self-driving laboratories to real-time bioprocessing
This report explores how data-driven systems improve reproducibility, speed decisions and make scale achievable across research and development.
Inside the report:
- Advance discovery through miniaturised, high-throughput and animal-free systems
- Integrate AI, robotics and analytics to speed decision-making
- Streamline cell therapy and bioprocess QC for scale and compliance
- And more!
This report unlocks perspectives that show how automation is changing the scale and quality of discovery. The result is faster insight, stronger data and better science – access your free copy today
Next-generation sequencing (NGS) technology has commonly been used to detect disease biomarkers. However, NGS has not proven to be successful at accurately detecting TERT promoter mutations because complexities in the DNA sequence in and around the TERT promoter make amplification by PCR extremely difficult. This limits the read depth of NGS and reduces its sensitivity in detecting mutations. While other detection methods such as quantitative PCR (qPCR) exist, they are not sensitive or precise enough for clinical applications such as monitoring cell-free DNA in plasma.
To overcome these challenges, Prof Polsky and his colleagues at NYU Langone Medical Centre and its Perlmutter Cancer Centre and scientists at Bio-Rad’s Digital Biology Group used ddPCR technology, which can reliably detect and quantify rare mutations with high sensitivity (<0.1 percent) independent of a reference standard.
The researchers developed two ddPCR-based assays to specifically detect the TERT C228T and C250T mutations in clinical samples. The limits of detection for the two assays were 0.062 and 0.051 percent.
In addition, the researchers evaluated the clinical application of the ddPCR assays by testing them in both formalin-fixed paraffin-embedded (FFPE) tissues and plasma samples from patients with metastatic melanoma. They found that the assays detected the mutation in 78 percent of samples and yielded no false positives. The test was able to detect mutation frequencies as low as 0.06 percent, demonstrating its high sensitivity.
“We have found the ddPCR workflow to be quite simple, and we estimate that the cost of the assays would be amenable to patient monitoring, where it would need to be repeated periodically as part of medical care,” said Prof Polsky.
The study was published in the Journal of Molecular Diagnostics.
Related topics
Analysis, Analytical Techniques, Biomarkers, Disease Research, DNA
Related conditions
Cancer
Related organisations
NYU Langone Medical Centre
Related people
Dr David Polsky