Study identifies novel measles drug target
Posted: 23 August 2019 | Victoria Rees (Drug Target Review) | No comments yet
Researchers have discovered the mechanism behind the viral genome for measles, which could serve as a druggable target.
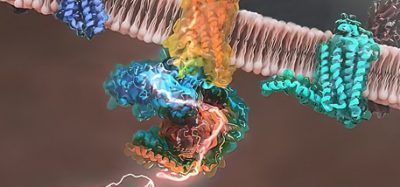
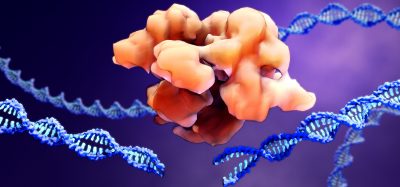
A new study has found that targeting specific areas of the measles virus polymerase, a protein complex that copies the viral genome, can effectively fight the measles virus. The researchers suggest that their findings can be used as an approach to develop new antiviral drugs.
The new research, from the Institute for Biomedical Sciences at Georgia State University, US, identified a novel protein interface in the polymerase complex vital for the regulation of polymerase activity.
“We have advanced current understanding of the underlying mechanism of viral RNA-dependent RNA polymerase advancement along the encapsidated genome – a poorly understood and not well characterised mechanism – by identifying and characterising the dynamic interactions between its constituents,” said Venice Du Pont, first author of the study.
The researchers found a novel model of dynamic binding and dissociation interacting proteins in the viral polymerase complex. The study discovered how the polymerase negotiates and regulates advancement along the viral genome. According to the researchers, this revealed a vulnerability that could be exploited as a target for small-molecule antiviral drugs.
“In recent years, enormous progress has been made to successfully target protein-protein interfaces for therapeutic purpose with chemical fragment libraries,” said Dr Richard Plemper, professor in the Institute for Biomedical Sciences and senior author on the study. “The newly identified interface in the paramyxovirus polymerase machinery meets key criteria of a druggable target and is very likely mechanistically conserved in highly pathogenic paramyxoviruses closely related to measles virus such as the zoonotic Nipah virus, against which we have currently no treatment option.”
The results were published in PLoS Pathogens.
Related topics
Drug Targets, Genomics, Research & Development
Related conditions
Measles, Nipah virus
Related organisations
Institute for Biomedical Sciences at Georgia State University, PLOS Pathogens
Related people
Dr Richard Plemper, Venice Du Pont