New AI software shows the nature of RNA-protein interactions
Posted: 31 October 2019 | Rachael Harper (Drug Target Review) | No comments yet
New software has been developed that can reveal the detailed RNA-binding properties of proteins, which is important for characterising the pathology of many diseases.
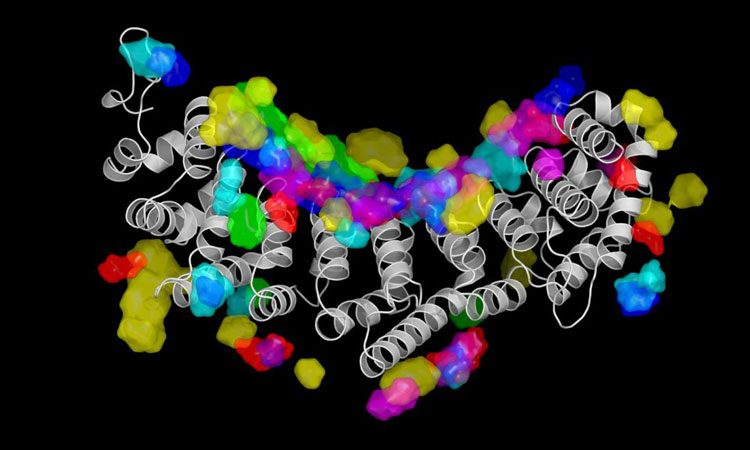

This depicts the RNA interaction surface of Fem-3-binding-factor 2 as predicted by NucleicNet (credit: 2019 KAUST).
A new computational tool called NucleicNet has been developed which uses artificial intelligence (AI) to infer the RNA-binding properties of proteins and provides additional biological insights that could aid in drug design and development.
The software was developed by scientists at King Abdullah University of Science and Technology (Kaust), Saudi Arabia.
“RNA-binding is a fundamental feature of many proteins,” said Jordy Homing Lam, a former research associate at KAUST and co-first author of the study. “Our structure-based computational framework can reveal the detailed RNA-binding properties of these proteins, which is important for characterising the pathology of many diseases.”
To better understand which parts of an RNA molecule tend to bind on different surface points of a protein, Lam and his colleagues turned to deep learning, a type of AI and taught NucleicNet to automatically learn the structural features that underpin interactions between proteins and RNA.
They trained the algorithm using three-dimensional structural data from 158 different protein-RNA complexes available on a public database. Pitting NucleicNet against other predictive models (all of which rely on sequence inputs rather than structural information) the team showed that the tool could most accurately detect which sites on a protein surface bound to RNA molecules or not.
What’s more, unlike any other model, NucleicNet could predict which aspects of the RNA molecule were doing the binding, be it part of the sugar-phosphate backbone or one of the four letters of the genetic alphabet.
In collaboration with researchers in China and the United States, the Kaust team validated their algorithm on a diverse set of RNA-binding proteins, including proteins implicated in gum cancer and amyotrophic lateral sclerosis, to show that the interactions deduced by NucleicNet closely matched those revealed by experimental techniques.
The research team reported their findings in Nature Communications.
Related topics
Artificial Intelligence, Big Data, Drug Development, Informatics, Protein, Research & Development, RNAs, Technology
Related organisations
King Abdullah University of Science and Technology (KAUST)
Related people
Jordy Homing Lam



