Technique for 2D printing of cells developed by researchers
Posted: 19 March 2020 | Victoria Rees (Drug Target Review) | No comments yet
A study has demonstrated the success of a 2D printing method to create cell arrays that reflect the body’s natural environment, allowing researchers to study stem cell differentiation.

Researchers report that they have created a new technique that can rapidly ‘print’ two-dimensional (2D) arrays of cells and proteins that mimic a wide variety of cellular environments in the body.
Conducted at the University of California (UC) Berkeley, US, the scientists used photolithography and programmable DNA to create their cellular formation.
By repeating the process, up to 10 different kinds of cells or proteins can be tethered to the surface in an arbitrary pattern”
According to the researchers, this technique could help scientists to develop a better understanding of the complex cell-to-cell messaging that dictates a cell’s final fate, from neural stem cell differentiating into a brain cell to a tumour cell with the potential to metastasise to an embryonic stem cell becoming an organ cell.
“What’s really powerful about this platform is you can create in vitro tissues that capture the spatial organisation of cells in the body, from the intestinal lining of your digestive tract to the arrangements of different cell types in the liver,” said Olivia Scheideler, who completed the research as a graduate student at Berkeley.
“Essentially, what this technique allows us to do is pattern different kinds of conditions in one shot and in a high-throughput manner,” said Lydia Sohn, Chancellor’s Professor of Mechanical Engineering at UC Berkeley and senior author of the paper. “It provides a whole range of options for what you could study, because it’s so flexible. You can pattern many different kinds of cells or proteins.”
A DNA tether
The researchers explain that in the new technique, each cell or protein is tethered to a substrate with a short string of DNA. It takes advantage of a patterning process called photolithography to attach, or print, each type of cell protein in one quick batch, greatly speeding up the process.
This works by exposing a coated surface or substrate to a pattern of light, which initiates a chemical reaction that dissolves the coating in the illuminated areas, leaving a templated substrate. The substrate is then bathed in strands of single-sided DNA, whose ends have been chemically altered to firmly latch on where the coating has been dissolved.
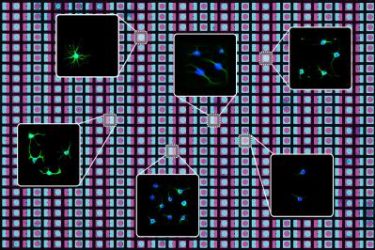

University of California Berkeley researchers have created a new technique that utilises photolithography and programmable DNA to rapidly ‘print’ two-dimensional arrays of cells and proteins that mimic a wide variety of cellular environments in the body [credit: UC Berkeley graphic by Olivia Scheideler].
Each single-sided DNA strand is programmed have a specific sequence of the nucleotides adenine (A), thymine (T), guanine (G) and cytosine (C). Single-sided DNA strands with the complementary nucleotide sequence are embedded or attached to cells or proteins of interest.
Finally, the surface is washed with a mixture of cells or proteins attached to the complementary strands of single-sided DNA, which bond with the single-sided DNA already attached to the surface to form double-helix ‘tethers’.
By repeating the process, up to 10 different kinds of cells or proteins can be tethered to the surface in an arbitrary pattern.
Scheideler used the new technique to pattern neural stem cells onto thousands of different arrays of two proteins, FGF-2 and ephrin-B2, to see how the spatial organisation of these two messenger signals helps determine the cells’ ultimate fate.
She found that many stem cells differentiated into mature neurons, even when they were primarily in contact with FGF-2, or ‘stay a stem cell’, messengers. On closer investigation, she found that those cells that differentiated were more likely to have small, finger-like extensions, or ‘neurites’, that touched the ephrin-B2 or ‘differentiate’ messengers.
“The great thing about this patterning technology is you can easily replicate these little patterns hundreds or thousands of times across a slide,” co-author David Schaffer said. “It is like running a thousand little independent experiments, each of which is a trial run to see how a stem cell listens to the cells around it. And then you can get very, very deep statistics about the various ways that it can be regulated.”
The study was published in Science Advances.
Related topics
Cell Cultures, Disease Research, Regenerative Medicine, Research & Development, Stem Cells
Related organisations
University of California Berkeley
Related people
David Schaffer, Lydia Sohn, Olivia Scheideler