Gramicidin A antibiotic analogues safe for human cells developed
Posted: 6 October 2020 | Victoria Rees (Drug Target Review) | No comments yet
A team has developed 10 new versions of the antibiotic gramicidin A, which they say should be safe for use in pills or injections.

A new approach has been utilised by scientists to transform gramicidin A, one of the world’s oldest antibiotics, into versions that – in preliminary lab tests – appear to be safer, stronger drugs. The research was conducted at the University of Tokyo, Japan.
Gramicidin A was originally discovered in soil bacteria and became the first commercially manufactured antibiotic in the early 1940s. It is prescribed today as a topical cream or drops, but it cannot be used as a pill or injection. Gramicidin A kills bacteria by punching itself through the cell membrane, allowing the cell to leak out and the surroundings to leak in through ion channels. However, these unregulated ion channels can have the same effect on human cells when gramicidin A is used inside the body.
About 350 artificial analogues of gramicidin A have been developed over the past 80 years, all of which have properties similar to the original and thus cannot be used in humans.
Now, the team designed and analysed over 4,000 gramicidin A artificial analogues, identifying those which are safe for humans.
According to the researchers, gramicidin A is a spiral of 15 amino acids, the building blocks of peptides, which are short proteins. They strategically selected six of those amino acids that could be altered without losing essential aspects of gramicidin A’s normal structure. Each of those six amino acids could be exchanged with four different amino acids to change how the peptide bonds together, leading to a total of 4,096 variations.
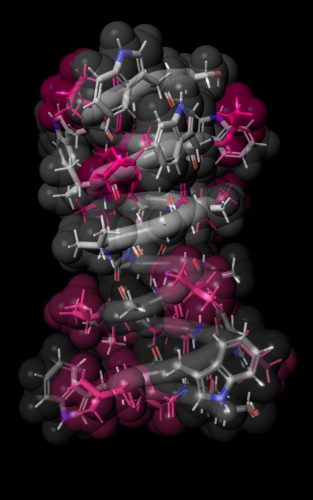

Pink colour highlights the locations of six amino acids that were altered in the synthesised versions of the gramicidin A molecule. A team from the University of Tokyo Department of Pharmaceutical Sciences has designed and analysed over 4,000 gramicidin A artificial analogues [credit: Hiroaki Itoh, first published in Nature Communications].
The one-bead-one-compound synthesis technique begins with small glass beads serving as the foundation to attach the first amino acid. The researchers built the peptide by attaching more amino acids one at a time. Whenever they reached the point of an amino acid variation, they split the beads into equal portions corresponding to the different amino acids, then remix the beads and continue building the peptide.
After completing their synthesis, the team placed each bead into its own container and analysed the function of their new versions of gramicidin A.
The researchers began testing their new versions of gramicidin A for activity against the common bacterial infection streptococcus. The strongest performers were then assessed for their potential ability to not indiscriminately kill human cells by testing their reactions with rabbit blood cells and mouse leukaemia cells.
These tests identified about 10 gramicidin A variations as promising future antibacterial drugs. The results also allowed the scientists to identify how specific structural changes to the amino acids affect the overall function of the molecule. They also measured the ion channel-forming ability of the best performing new versions of gramicidin A. Although they had reduced toxicity to mammalian cells, their ion channel-forming ability remained strong. Therefore, these subtle modifications to a few amino acids could transform gramicidin A’s ion channel-forming function from indiscriminate to bacteria specific.
“Most important is that this strategy can be used for other types of natural products and other ion channel-forming compounds. It has long been believed to be very difficult to realize species-selective ion channel-forming activity, but our study showed gramicidin A can have very bacteria-selective activity. I believe this thought can change the standard of ion channel-forming natural products,” said Assistant Professor Hiroaki Itoh, one of the authors of the research.
The study was published in Nature Communications.
Related topics
Antibiotics, Drug Development, Formulation, Ion Channels, Research & Development
Related organisations
University of Tokyo
Related people
Assistant Professor Hiroaki Itoh