Behavioural screen in C. elegans reveals Parkinson’s drug candidates
Posted: 24 February 2021 | Victoria Rees (Drug Target Review) | No comments yet
Researchers have developed a workflow to study the behaviour of C. elegans in a high-throughput screen to identify Parkinson’s disease drugs.
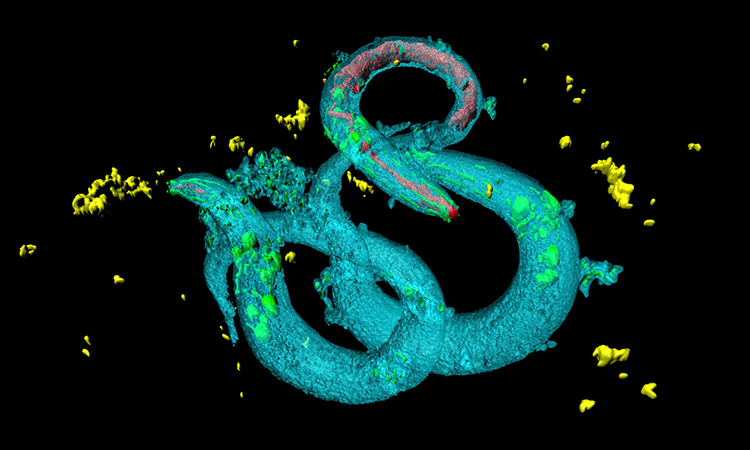

Researchers have developed a machine-based learning workflow that could be used in the discovery of new therapeutics for Parkinson’s disease. The study was conducted at Princeton University, US.
According to the researchers, they linked branched-chain amino acid transferase 1 (BCAT1) dysfunction with the movement disorder Parkinson’s disease and found that RNAi-mediated knockdown of neuronal bcat-1 in C. elegans causes abnormal spasm-like ‘curling’ behaviour with age.
The team say that the curling is similar to Parkinson’s disease motor symptoms, being both age-dependent and progressive. Due to this, they performed curling tests on day eight of adulthood.
Biomarkers aren’t just supporting drug discovery – they’re driving it
FREE market report
From smarter trials to faster insights, this report unpacks the science, strategy and real-world impact behind the next generation of precision therapies.
What you’ll unlock:
- How biomarkers are guiding dose selection and early efficacy decisions in complex trials
- Why multi-omics, liquid biopsy and digital tools are redefining the discovery process
- What makes lab data regulatory-ready and why alignment matters from day one
Explore how biomarkers are shaping early drug development
Access the full report – it’s free!
To simplify quantification and maintain a low data overhead, the team say that their simple segment-train-quantify platform enables fully automated scoring of image stills upon training of a convolutional neural network. They trained a neural network for the detection and classification of worm postures in order to carry out high-throughput curling analysis without the need for user intervention or post-inspection.
In a proof-of-concept screen, the researchers tested 50 US Food and Drug Administration (FDA)-approved drugs: enasidenib, ethosuximide, metformin and nitisinone. To carry out a manual thrashing assay, 100 worms per treatment group were individually picked and 30 second videos of around 10 worms each are recorded. The researchers found that the drugs were identified as candidates for potential late-in-life intervention in Parkinson’s disease.
The scientists say that these findings point to the utility of their high-throughput platform for the automated recording of worm postures and in particular, the discovery of potential candidate treatments for Parkinson’s disease.
“The ability to perform large-scale screening for modifiers of Parkinson’s disease-like curling dysfunction in C. elegans may uncover novel disease mechanisms and identify potential therapeutics,” the authors of the paper write.
The findings from the study were published in Communications Biology.
Related topics
Drug Development, Drug Discovery, Drug Repurposing, High-Throughput Screening (HTS), Screening
Related conditions
Parkinson's disease
Related organisations
Princeton University