Antibody characterisation – an essential researchers’ resource
Posted: 13 December 2019 | Nikki Withers (Drug Target Review) | No comments yet
Central to reproducibility in biomedical research is the ability to use well-characterised and defined reagents. The CPTAC Antibody Portal serves as a National Cancer Institute community resource that provides access to many standardised renewable affinity reagents to cancer-associated targets and accompanying characterisation data. Nikki Withers spoke to Dr Tara Hiltke who oversees the NCI’s Antibody Characterization Program to hear how researchers can benefit from this valuable resource.
ANTIBODIES are among the most commonly used tools in the biological sciences and are utilised in numerous experiments to identify and isolate other molecules. However, many cancer researchers lack access to affordable, well-characterised and analytically validated renewable affinity reagents, a problem that could be hindering cancer biomarker discovery and validation, cancer diagnostics development and therapeutics monitoring.1 In an effort to accelerate cancer research and provide well-characterised monoclonal antibodies to the scientific community, the National Cancer Institute’s (NCI’s) Office of Cancer Clinical Proteomics Research launched the Antibody Characterization Program as part of the Clinical Proteomic Technologies for Cancer (CPTC) initiative. “This is a community resource for scientists based on what they need,” explained Program Director, Dr Tara Hiltke. “Our reagents are produced, screened and selected for specific applications, so it is possible for us to work with researchers to create antibodies that could potentially be used for therapeutic applications.”
A much-needed resource
Fundamental to the development of the programme was the NCI’s Proteomic Technologies Reagents Resource Workshop, which took place in 2005. The aim of the workshop was to identify the cancer research community’s expressed needs for validated and well-characterised affinity capture reagents (eg, antibodies, aptamers and affibodies) to advance proteomics research platforms for the prevention, early detection, treatment and monitoring of cancer. The workshop involved leading scientists in proteomics research, who discussed model systems for evaluating and delivering affinity reagents to the research community to support proteomics-based research.
Following the workshop, a report2 was published, which highlighted the many challenges and opportunities for developing a community-wide resource. “The current challenges that cancer investigators face with respect to affinity capture reagents may be illustrated by the case of monoclonal antibodies, currently the most mature affinity capture methodology,” states the report. “Numerous antibodies and commercial pipelines for antibody production are currently in place. However, the majority of antibodies are poorly characterised and not adequately validated for the variety of applications of interest to the research community (Table 1). As such, the user must navigate through an increasingly complex marketplace to determine whether data are available on the binding characteristics of an antibody and whether the antibody is suitable for a specific application.”2
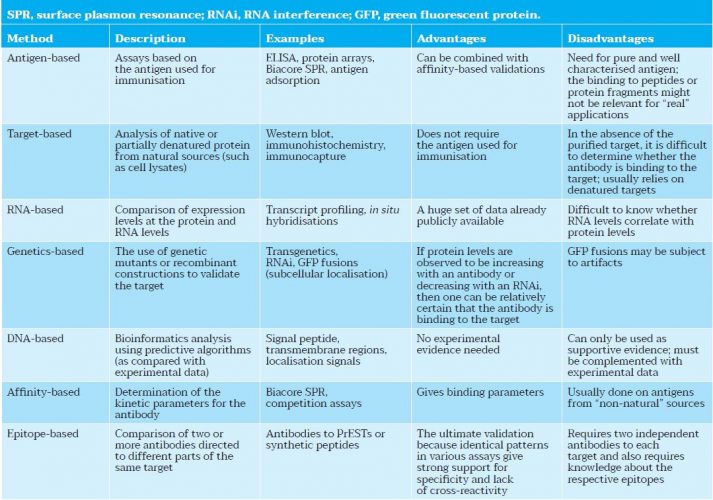
Table 1: Application-independent validation methods for antibodies adapted from Uhlen et al.3)
For example, the report states that a search of Biocompare – an online search tool for scientific products and resources – for monoclonal antibodies to p53 returned more than 1,300 choices representing more than 50 vendors. These antibodies varied widely in application, design controls, validation parameters, supporting documentation and cost. Conversely, a search for emerging candidate biomarkers, such as CA27.29 – a candidate biomarker for breast cancer – yielded no results. Additionally, a query for CA15.3 identified several vendors with available monoclonal antibodies to this candidate breast cancer marker, whereas a syntactical change to CA15.3 returned no results.
This example, according to the report, identifies several needs of the cancer research community. Those requirements are outlined here, alongside their anticipated benefits:
- A set of well characterised and validated capture reagents with readily accessible supporting data will conserve resources by enabling investigators to rapidly determine reagent suitability for specific applications.
- Identifying the gaps in reagent needs will support the development of well characterised antibodies to prospective and emerging targets for which there is no established commercial market.
- Appropriate annotations and ontologies will be needed for each of these resources to accommodate the variety of search terms used to query for the same resource and enable researchers to locate the reagents that best meet the needs of their research projects.
- Streamlined, high-throughput methods of reagent preparation can improve the cost efficiency of production and characterisation, thereby resulting in the development of advanced proteomics analysis platforms.
The report concludes that a centralised, shared reagents resource will “serve the research community most effectively by coordinating and maintaining an open access database of primary characterisation and validation data for new and existing affinity reagents.”
What is the Antibody Characterization Program?
Many cancer researchers lack access to affordable, well-characterised and analytically validated renewable affinity reagents, a problem that could be hindering cancer biomarker discovery and validation”
The goal of the NCI’s Antibody Characterization Program is to have up to three monoclonal antibodies produced for each successfully expressed/purified recombinant antigen and one antibody per peptide (one to three peptides per protein). To date, over 5,000 clones have been screened before selecting the current 583 antibodies. They are winnowed down based on the projected end use of the antibody. The programme typically selects up to three monoclonal antibodies that are further extensively characterised at the NCI Antibody Characterisation Laboratory (ACL) within the Frederick National Laboratory for Cancer Research by indirect ELISA, IP-MS, western blot, IHC, affinity measurement (ie, SPR) and Nucleic Acid Programmable Protein Array (NAPPA) (described in the pipeline below) and all data is made available to the scientific community here.
Explaining how the process works, Dr Hiltke said: “Scientists can submit their application and, in this application, they have to provide information on the target and its intended use. The applicant must provide the protein or peptide used for the immunisation, screening and characterisation; we’re not just accepting target names, we need the material to start the process. After that, we produce the antibodies and work with the applicant to screen and select the appropriate antibody for their research method. Ultimately, all the antibodies are also characterised in the ACL to provide more details on these antibodies using standard assay methods.”
 To be accepted into the programme, the target must be cancer related and, if there are other antibodies available to the target, the applicant must show they have a unique method or that all other antibodies available are not working. “We don’t want to make another antibody to something that there’s already 500 antibodies available to,” says Dr Hiltke.
To be accepted into the programme, the target must be cancer related and, if there are other antibodies available to the target, the applicant must show they have a unique method or that all other antibodies available are not working. “We don’t want to make another antibody to something that there’s already 500 antibodies available to,” says Dr Hiltke.
All applications are reviewed by an independent committee and requests are prioritised. For the application to be responsive, there are several things the applicant must agree on. Firstly, all reagents go to the Developmental Studies Hybridoma Bank (DSHB) where they are sold for research use only. Directed by David R Soll, the DSHB was established by the National Institutes of Health in 1986 to supply research investigators worldwide with low cost monoclonal antibodies for studies in developmental and cell biology. The DSHB has continued to expand its collection through contributions from researchers and now offers over 3,500 monoclonal antibodies. The second is that all antibodies are labelled with the prefix CPTC– Clinical Proteomic Technologies for Cancer. Whenever antibodies created from the programme are referenced, this prefix must be used in the publication. If third-party commercialisation occurs, the team must use this prefix so that other researchers can track it back to the programme. “This is one of the challenges with antibodies; often more than one vendor will sell the same antibody,” says Dr Hiltke. “We hope that adding the CPTC reference for all antibodies created through our programme will help prevent people from buying the same antibody multiple times and wasting their time and money on the same thing.”
Researchers in the programme are encouraged to participate in the screening and selection process”
One of the questions Dr Hiltke is often asked is “How does the research scientist benefit from this?” She explained: “The main thing is that the applicant is getting a fit-for-purpose antibody, screened and selected for a specific and intended purpose, for free to use in their research while the scientific community gets a low cost, well characterised antibody through DSHB. We want to make sure that while we’re screening and doing the selections, we are focusing on the antibodies that are likely to work for the intended purpose or method. Researchers in the programme are encouraged to participate in the screening and selection process; we aren’t working in silo, we are working closely together. The goal is to select the antibodies for their intended application in a fit-for-purpose way. They will receive a clone at the end that can be used for internal research and/or commercial collaborations.”
CPTC antibody characterisation pipeline
In order to produce antibodies with consistent quality, the programme uses a standardised antibody characterisation pipeline to ensure rigour and reproducibility. Using a team of experts, the following pipeline is executed for each successful antibody candidate:
- Antigen selection – approved target antigens are provided by investigators and QC tested by ACL
- Antibody production – monoclonal antibodies are produced, typically in mice or rabbits
- Antibody initial screening – up to 30 clones per antibody are screened using methods based on end-use applications. Typically, up to three clones are chosen for final antibody production.
- Antibody characterisation laboratory – antibody isotype, SDS-PAGE, western blot, indirect ELISA, affinity measurement such as surface plasma resonance, immunohistochemistry, immuno-mass spectroscopy, NAPPA. All data (positive and negative) is made available at the NCI Antibody Portal.
- Distribution and community access – DSHB at the University of Iowa.
Dr Hiltke concludes that what makes the programme unique is its fit-for-purpose approach. “I really want people to understand that this is a great resource and they should take advantage of it. Currently there is an open application for antibody production target requests now through 13 February 2020 here.
Further information
To find out more about the programme, visit here.
 Dr Tara Hiltke provides leadership and oversight to the Monoclonal Antibody Characterization Program. She also works in developing other methods of antigen generation. Previously, Tara served as a senior scientist/ project manager in assay development at both Wellstat Diagnostics and BioVeris Corporation, where she developed clinical assays for diagnostic markers using the electro-chemiluminescence platform and magnetic beads. Tara holds a PhD degree (1999) in biology from the University of Buffalo.
Dr Tara Hiltke provides leadership and oversight to the Monoclonal Antibody Characterization Program. She also works in developing other methods of antigen generation. Previously, Tara served as a senior scientist/ project manager in assay development at both Wellstat Diagnostics and BioVeris Corporation, where she developed clinical assays for diagnostic markers using the electro-chemiluminescence platform and magnetic beads. Tara holds a PhD degree (1999) in biology from the University of Buffalo.
References
- [Internet]. Aacc.org. 2019 [cited 2 December 2019]. Available from: https://www.aacc.org/~/media/files/divisions/ncicallforantibodytargets.pdf?la=en
- Haab B, Paulovich A, Anderson N, Clark A, Downing G, Hermjakob H et al. A Reagent Resource to Identify Proteins and Peptides of Interest for the Cancer Community: Molecular & Cellular Proteomics. 2006;5(10):1996-2007.
- Uhlen M et al. A human protein atlas for normal and cancer tissues based on antibody proteomics. Cell. Proteomics. 2005;4:1920-1932
Related topics
Antibodies, Antibody Discovery, Proteomics, Research & Development
Related conditions
Cancer
Related organisations
Biocompare, Clinical Proteomic Technologies for Cancer (CPTC), Developmental Studies Hybridoma Bank (DSHB), Frederick National Laboratory for Cancer Research, National Cancer Institute (NCI), National Institutes of Health, NCI Antibody Characterisation Laboratory (ACL), NCI Office of Cancer Clinical Proteomics Research
Related people
Dr Tara Hiltke (National Cancer Institute)