Newly discovered protein could be used to produce life-saving antifungals
Posted: 29 September 2022 | Ria Kakkad (Drug Target Review) | No comments yet
Researchers have discovered a protein, Ait1, that is responsible for controlling cell growth in yeasts. Since humans and yeasts have remarkably similar cellular mechanisms, teasing out the differences presents drug developers with new target.

Yeast cells and human immune system cells rely on surprisingly similar chemical reactions to know when to grow. University of Arizona, US, scientists have identified subtle differences between the two cell types that could help spur the development of antifungal drugs that are able to attack disease-causing yeasts in the body while sparing the immune system. The team also discovered a protein, Ait1, that is responsible for controlling cell growth in yeasts.
Their findings, published in eLife, not only have implications for drug development, they also provide important insight into the evolution of an ancient growth control pathway found in all multicellular organisms.
It is well known in the scientific community that a conglomerate of proteins called Target of Rapamycin kinase Complex 1 (TORC1) controls the growth of cells in everything from humans to yeasts. But researchers have now identified and named the protein that triggers this process in yeasts – a nutrient sensor and TORC1 regulator they named Ait1. When working normally, Ait1 shuts down TORC1 in yeasts when cells are starved for nutrients, blocking cell growth.
The researchers were interested in determining how cells sense stress and starvation and then decide how fast to grow. Understanding how TORC1 is triggered in different organisms is important for developing treatments for a wide variety of diseases.
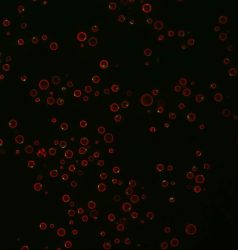

Yeast with a red fluorescent protein marking the vacuole – the nutrient storage compartment of the cell – and a green fluorescent protein marking aggregates of TORC1 that form in cells missing Ait1
[Credit: Andrew Capaldi and team].
TORC1 was originally discovered in yeast, but it is also critical for the activation of cells in the human immune system to mount a response. When TORC1 is notworking as it should, it can trigger the development of cancer, diabetes and various neurological disorders including epilepsy and depression.
But the fact that human bodies depend on the same TORC1 pathway as yeast presents a problem.
“As an example, you can block the growth of yeast very easily using rapamycin – a drug that binds directly to and inhibits TORC1 – so that would fight any infection well,” explained Associate Professor Andrew Capaldi, the study’s co-author. “However, that very same drug is regularly used in transplant patients to suppress their immune system, so that would be a disaster.”
The researchers found that while the TORC1 pathway is very similar in yeast and humans, humans don not rely on Ait1 to regulate TORC1. So, drugs that specifically target Ait1 should inhibit the growth of yeast and not human immune cells.
Ait1 has only evolved in the last 200 million years, which is relatively recent in evolutionary terms. About 200 million years ago a TORC1 regulator called Rheb seems to have disappeared from the cells of various organisms exactly when Ait1 evolved.
“We showed that some of the ancient TORC1 regulators found in humans (including Rheb) have been lost in the same yeasts that gained Ait1 200 million years ago,” Capaldi concluded. “These same ancient regulators have also been lost in the evolution of other single-celled organisms, including many parasites and plants. So, it is very likely that other single-celled organisms gained new regulators – similar to Ait1 – of their own. Now people can go out and look for them, as they will also be good drug targets.”
Related topics
Molecular Biology, Molecular Targets, Protein
Related conditions
Cancer, Diabetes, Neurological disorders
Related organisations
University of Arizona
Related people
Associate Professor Andrew Capaldi