Enzyme involved in cancer growth imaged for first time
Posted: 7 November 2019 | Victoria Rees (Drug Target Review) | No comments yet
A study has revealed the structure of an enzyme that promotes cancer growth by mutating RNA, which could be used in the development of cancer treatments.

Researchers have discovered the structure of a novel RNA-modifying enzyme and identified the mechanism that controls how it recognises its substrate. According to the team, their findings could be used to inform the development of therapeutics for cancer.
The study was conducted at the University of California, Riverside, US.
The researchers investigated the ZCCHC4 enzyme, which influences cell proliferation and is linked to cancers. It does this by introducing an RNA modification, N6-methyladenosine (m6A), into ribosomes.
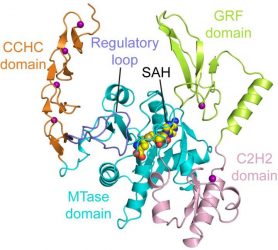

This image shows the structure of ZCCHC4 (credit: Song lab, UC Riverside).
ZCCHC4 is overexpressed in tumours associated with hepatocellular carcinoma, the most common type of primary liver cancer.
To discover the structure of ZCCHC4, the researchers produced an enzymatically active and structurally rigid ZCCHC4 fragment. They then coaxed this to crystallise and finally diffracted the crystals using X-rays. Analysing this data enabled them to structure the enzyme.
“This is the first time anyone has determined the crystal structure of ZCCHC4,” lead researcher associate professor Jikui Song said. “Our discovery can be used for structure-based drug design against cancers and lead to a better understanding of how m6A, a modification associated with numerous biological processes, is installed on ribosomal RNA.”
Due to the new insight into ZCCHC4’s structure, the researchers were also able to determine how the enzyme is built to specifically act on 28S ribosomal RNA.
“We now understand that this enzyme is controlled by an ‘autoinhibitory’ mechanism that has been observed in many other cellular processes,” continued Song.
The results were published in Nature Communications.
Related topics
DNA, Drug Targets, Enzymes, Research & Development, RNAs
Related conditions
Cancer
Related organisations
University of California Riverside
Related people
Jikui Song